from katlas.core import *
import pandas as pdBackground frequencies
df = Data.get_ks_unique()PSSMs - ks dataset
def get_bg_dict(df, acceptor, seq_col='site_seq'):
site = df[df.acceptor.isin(list(acceptor))].copy()
site_pssm = get_prob(site, seq_col)
return flatten_pssm(site_pssm, True)acceptors = ['S','T','Y','ST','STY']index_names = 'ks_' + pd.Series(acceptors)bg_pssms = pd.DataFrame([get_bg_dict(df,acceptor) for acceptor in acceptors],index=index_names)bg_pssms| -20P | -20G | -20A | -20C | -20S | -20T | -20V | -20I | -20L | -20M | ... | 20H | 20K | 20R | 20Q | 20N | 20D | 20E | 20s | 20t | 20y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ks_S | 0.07430 | 0.06810 | 0.07593 | 0.01466 | 0.05325 | 0.03852 | 0.05129 | 0.04034 | 0.07899 | 0.02027 | ... | 0.02129 | 0.07033 | 0.05931 | 0.04420 | 0.03627 | 0.05488 | 0.07732 | 0.04346 | 0.01498 | 0.00645 |
| ks_T | 0.06163 | 0.06590 | 0.07427 | 0.01644 | 0.04383 | 0.03439 | 0.05966 | 0.04018 | 0.08948 | 0.02481 | ... | 0.02239 | 0.07507 | 0.06056 | 0.04526 | 0.03801 | 0.05867 | 0.07664 | 0.03548 | 0.01987 | 0.00899 |
| ks_Y | 0.05618 | 0.07242 | 0.07067 | 0.01595 | 0.04448 | 0.03877 | 0.05589 | 0.04594 | 0.08632 | 0.02429 | ... | 0.01950 | 0.07115 | 0.05789 | 0.04250 | 0.03763 | 0.05622 | 0.08318 | 0.02316 | 0.01249 | 0.01737 |
| ks_ST | 0.07050 | 0.06744 | 0.07543 | 0.01520 | 0.05042 | 0.03728 | 0.05380 | 0.04029 | 0.08214 | 0.02163 | ... | 0.02162 | 0.07174 | 0.05969 | 0.04452 | 0.03679 | 0.05601 | 0.07712 | 0.04108 | 0.01644 | 0.00721 |
| ks_STY | 0.06710 | 0.06863 | 0.07430 | 0.01537 | 0.04901 | 0.03763 | 0.05430 | 0.04163 | 0.08313 | 0.02226 | ... | 0.02112 | 0.07160 | 0.05926 | 0.04404 | 0.03699 | 0.05606 | 0.07855 | 0.03685 | 0.01551 | 0.00961 |
5 rows × 943 columns
def get_site_cnt(df, acceptor, seq_col='site_seq'):
site = df[df.acceptor.isin(list(acceptor))].copy()
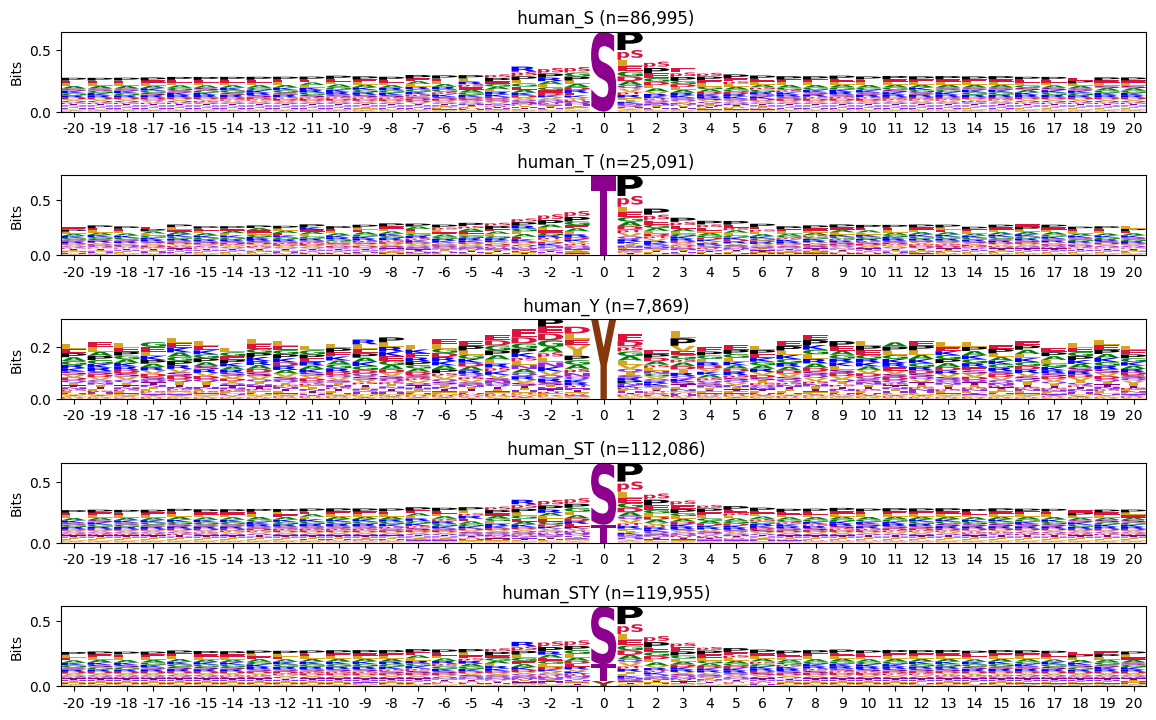
return len(site)cnt = {name:get_site_cnt(df,acceptor) for acceptor,name in zip(acceptors,index_names)}plot_logos(bg_pssms,cnt,prefix='')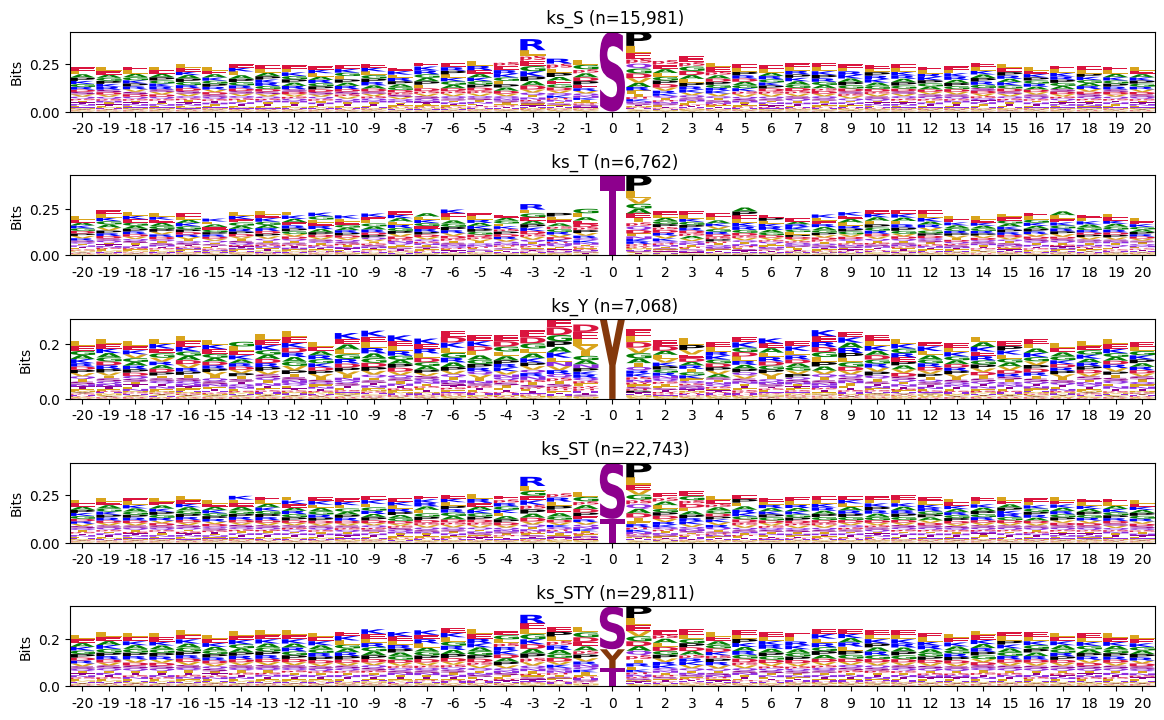
PSSMs - human phosphoproteome
human = Data.get_human_site()human['acceptor']=human.site.str[0]human.head(1)| substrate_uniprot | substrate_genes | site | source | AM_pathogenicity | substrate_sequence | substrate_species | sub_site | substrate_phosphoseq | position | site_seq | acceptor | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | A0A024R4G9 | C19orf48 MGC13170 hCG_2008493 | S20 | psp | NaN | MTVLEAVLEIQAITGSRLLSMVPGPARPPGSCWDPTQCTRTWLLSH... | Homo sapiens (Human) | A0A024R4G9_S20 | MTVLEAVLEIQAITGSRLLsMVPGPARPPGSCWDPTQCTRTWLLSH... | 20 | _MTVLEAVLEIQAITGSRLLsMVPGPARPPGSCWDPTQCTR | S |
index_names2 = 'human_' + pd.Series(acceptors)bg_pssms_human = pd.DataFrame([get_bg_dict(human,acceptor) for acceptor in acceptors],index=index_names2)bg_pssms_human| -20P | -20G | -20A | -20C | -20S | -20T | -20V | -20I | -20L | -20M | ... | 20H | 20K | 20R | 20Q | 20N | 20D | 20E | 20s | 20t | 20y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| human_S | 0.08364 | 0.06732 | 0.07243 | 0.01298 | 0.06843 | 0.04076 | 0.04860 | 0.03262 | 0.08046 | 0.01853 | ... | 0.02240 | 0.06643 | 0.06452 | 0.04737 | 0.03316 | 0.05179 | 0.07982 | 0.04460 | 0.01317 | 0.00367 |
| human_T | 0.07986 | 0.06436 | 0.07476 | 0.01194 | 0.06270 | 0.04093 | 0.05299 | 0.03346 | 0.07854 | 0.02181 | ... | 0.02007 | 0.07009 | 0.06264 | 0.04805 | 0.03220 | 0.05181 | 0.07866 | 0.04366 | 0.01702 | 0.00485 |
| human_Y | 0.07390 | 0.06714 | 0.07310 | 0.01602 | 0.05840 | 0.03946 | 0.05178 | 0.04145 | 0.08741 | 0.02026 | ... | 0.02177 | 0.06572 | 0.05960 | 0.04994 | 0.03130 | 0.05538 | 0.07811 | 0.03470 | 0.01619 | 0.01660 |
| human_ST | 0.08279 | 0.06666 | 0.07295 | 0.01275 | 0.06714 | 0.04079 | 0.04959 | 0.03281 | 0.08003 | 0.01927 | ... | 0.02188 | 0.06725 | 0.06410 | 0.04752 | 0.03295 | 0.05180 | 0.07956 | 0.04439 | 0.01403 | 0.00394 |
| human_STY | 0.08221 | 0.06669 | 0.07296 | 0.01296 | 0.06657 | 0.04071 | 0.04973 | 0.03338 | 0.08052 | 0.01933 | ... | 0.02187 | 0.06715 | 0.06381 | 0.04768 | 0.03284 | 0.05203 | 0.07946 | 0.04377 | 0.01417 | 0.00475 |
5 rows × 943 columns
all_pssms = pd.concat([bg_pssms,bg_pssms_human])# all_pssms.to_parquet('~/katlas/dataset/CDDM/ks_background.parquet')Accessible through Data.get_ks_background
cnt_human = {name:get_site_cnt(human,acceptor) for acceptor,name in zip(acceptors,index_names2)}cnt.update(cnt_human)cnt{'ks_S': 15981,
'ks_T': 6762,
'ks_Y': 7068,
'ks_ST': 22743,
'ks_STY': 29811,
'human_S': 86995,
'human_T': 25091,
'human_Y': 7869,
'human_ST': 112086,
'human_STY': 119955}plot_logos(bg_pssms_human,cnt,prefix='')