cut_seq('AAkUuPSFSTtH',-5,4)'AkUuPSFSTt'cut_seq (input_string:str, min_position:int, max_position:int)
Extract sequence based on a range relative to its center position
| Type | Details | |
|---|---|---|
| input_string | str | site sequence |
| min_position | int | minimum position relative to its center |
| max_position | int | maximum position relative to its center |
STY2sty (input_string:str)
Replace all ‘STY’ with ‘sty’ in a sequence
get_dict (input_string:str)
Get a dictionary of input string; no need for the star in the middle; make sure it is 15 or 10 length
| Type | Details | |
|---|---|---|
| input_string | str | phosphorylation site sequence |
multiply_func (values, factor=17)
Multiply the possibilities of the amino acids at each position in a phosphorylation site
| Type | Default | Details | |
|---|---|---|---|
| values | list of values, possibilities of amino acids at certain positions | ||
| factor | int | 17 | scale factor |
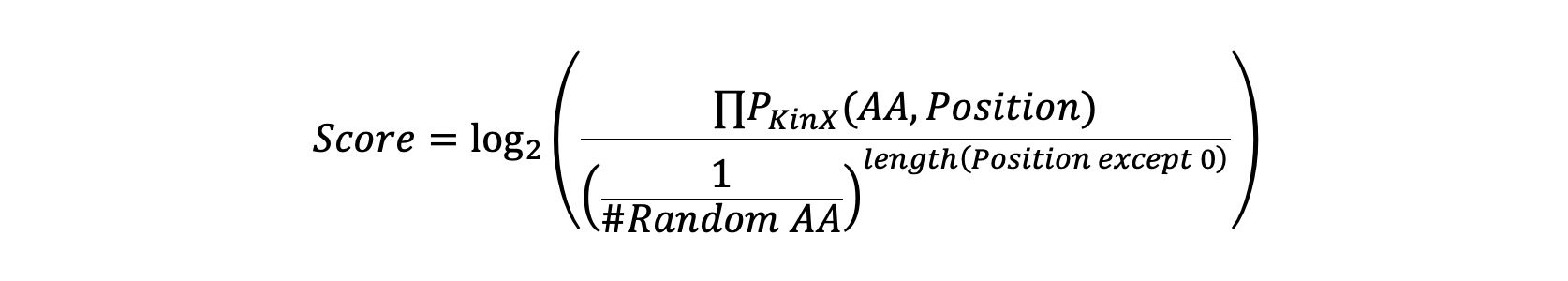
The function implement formula from Johnson et al. Nature: An atlas of substrate specificities for the human serine/threonine kinome, Supplementary Note2 (page 160)
Multiply class, consider the dynamics of scale factor
multiply (values, kinase, num_dict={'SYK': 18, 'PTK2': 18, 'ZAP70': 18, 'ERBB2': 18, 'CSK': 18, 'FGFR4': 18, 'EGFR': 18, 'ERBB4': 18, 'EPHA8': 18, 'EPHA7': 18, 'EPHA5': 18, 'EPHA2': 18, 'EPHB2': 18, 'EPHB1': 18, 'EPHB3': 18, 'EPHB4': 18, 'EPHA4': 18, 'EPHA3': 18, 'EPHA6': 18, 'FRK': 18, 'EPHA1': 18, 'TEC': 18, 'BTK': 18, 'ITK': 18, 'BMX': 18, 'TXK': 16, 'ABL2': 18, 'ABL1': 18, 'SRMS': 18, 'PTK2B': 18, 'FER': 18, 'MERTK': 18, 'AXL': 18, 'FES': 18, 'PTK6': 18, 'YES1': 18, 'FGR': 18, 'SRC': 18, 'FYN': 18, 'LCK': 18, 'BLK': 18, 'LYN': 18, 'HCK': 18, 'PDGFRB': 18, 'PDGFRA': 18, 'FLT3': 18, 'TYRO3': 18, 'ROS1': 18, 'TEK': 18, 'LTK': 18, 'ALK': 18, 'MUSK': 18, 'KIT': 18, 'CSF1R': 18, 'MET': 18, 'KDR': 18, 'RET': 18, 'MST1R': 16, 'JAK3': 16, 'FLT1': 16, 'MATK': 18, 'FGFR3': 18, 'FGFR2': 18, 'FGFR1': 18, 'FLT4': 18, 'INSR': 18, 'IGF1R': 18, 'INSRR': 16, 'NTRK3': 18, 'NTRK1': 18, 'NTRK2': 18, 'TNK1': 18, 'TNK2': 18, 'DDR2': 18, 'DDR1': 18, 'TYK2': 18, 'JAK2': 18, 'JAK1': 18, 'TNNI3K_TYR': 18, 'NEK10_TYR': 16, 'PINK1_TYR': 16, 'MAP2K7_TYR': 16, 'PKMYT1_TYR': 16, 'TESK1_TYR': 16, 'LIMK1_TYR': 16, 'LIMK2_TYR': 16, 'WEE1_TYR': 18, 'MAP2K6_TYR': 16, 'MAP2K4_TYR': 16, 'PDHK1_TYR': 16, 'BMPR2_TYR': 16, 'PDHK4_TYR': 16, 'PDHK3_TYR': 16, 'AAK1': 17, 'ACVR2A': 17, 'ACVR2B': 17, 'AKT1': 17, 'AKT2': 17, 'AKT3': 17, 'ALK2': 17, 'ALK4': 17, 'ALPHAK3': 17, 'AMPKA1': 17, 'AMPKA2': 17, 'ANKRD3': 17, 'ATM': 17, 'ATR': 17, 'AURA': 17, 'AURB': 17, 'AURC': 17, 'GRK2': 17, 'GRK3': 17, 'BCKDK': 17, 'BIKE': 17, 'BMPR1A': 17, 'BMPR1B': 17, 'BMPR2': 17, 'BRAF': 17, 'BRSK1': 17, 'BRSK2': 17, 'BUB1': 17, 'CAMK1A': 17, 'CAMK1B': 17, 'CAMK1D': 17, 'CAMK1G': 17, 'CAMK2A': 17, 'CAMK2B': 17, 'CAMK2D': 17, 'CAMK2G': 17, 'CAMK4': 17, 'CAMKK1': 17, 'CAMKK2': 17, 'CAMLCK': 17, 'CDK1': 17, 'CDC7': 17, 'CDK10': 17, 'CDK19': 17, 'CDK2': 17, 'CDK3': 17, 'CDK4': 17, 'CDK5': 17, 'CDK6': 17, 'CDK7': 17, 'CDK8': 17, 'CDK9': 17, 'CDKL1': 17, 'CDKL5': 17, 'CHAK1': 17, 'CHAK2': 17, 'CDK13': 17, 'CHK1': 17, 'CHK2': 17, 'CK1A': 17, 'CK1A2': 17, 'CK1D': 17, 'CK1E': 17, 'CK1G1': 17, 'CK1G2': 17, 'CK1G3': 17, 'CK2A1': 17, 'CK2A2': 17, 'CLK1': 17, 'CLK2': 17, 'CLK3': 17, 'CLK4': 17, 'COT': 17, 'CRIK': 17, 'CDK12': 17, 'DAPK1': 17, 'DAPK2': 17, 'DAPK3': 17, 'DCAMKL1': 17, 'DCAMKL2': 17, 'DLK': 17, 'DMPK1': 17, 'DNAPK': 17, 'DRAK1': 17, 'DYRK1A': 17, 'DYRK1B': 17, 'DYRK2': 17, 'DYRK3': 17, 'DYRK4': 17, 'ERK1': 17, 'ERK2': 17, 'ERK5': 17, 'ERK7': 17, 'MTOR': 17, 'GAK': 17, 'GCK': 17, 'GCN2': 17, 'GRK4': 17, 'GRK5': 17, 'GRK6': 17, 'GRK7': 17, 'GSK3A': 17, 'GSK3B': 17, 'HASPIN': 17, 'HGK': 17, 'HIPK1': 17, 'HIPK2': 17, 'HIPK3': 17, 'HIPK4': 17, 'HPK1': 17, 'HRI': 17, 'HUNK': 17, 'ICK': 17, 'IKKA': 17, 'IKKB': 17, 'IKKE': 17, 'IRAK1': 17, 'IRAK4': 17, 'IRE1': 17, 'IRE2': 17, 'JNK1': 17, 'JNK2': 17, 'JNK3': 17, 'KHS1': 17, 'KHS2': 17, 'KIS': 17, 'LATS1': 17, 'LATS2': 17, 'LKB1': 17, 'LOK': 17, 'LRRK2': 17, 'MAK': 17, 'MEK1': 17, 'MEK2': 17, 'MEK5': 17, 'MEKK1': 17, 'YSK4': 17, 'MEKK2': 17, 'MEKK3': 17, 'ASK1': 17, 'MEKK6': 17, 'MAP3K15': 17, 'MAPKAPK2': 17, 'MAPKAPK3': 17, 'MAPKAPK5': 17, 'MARK1': 17, 'MARK2': 17, 'MARK3': 17, 'MARK4': 17, 'MASTL': 17, 'MELK': 17, 'MINK': 17, 'MLK1': 17, 'MLK2': 17, 'MLK3': 17, 'MLK4': 17, 'MNK1': 17, 'MNK2': 17, 'MOK': 17, 'MOS': 17, 'MPSK1': 17, 'MRCKA': 17, 'MRCKB': 17, 'MSK1': 17, 'MSK2': 17, 'SRPK3': 17, 'MST1': 17, 'MST2': 17, 'MST3': 17, 'MST4': 17, 'MYO3A': 17, 'MYO3B': 17, 'NDR1': 17, 'NDR2': 17, 'NEK1': 17, 'NEK11': 17, 'NEK2': 17, 'NEK3': 17, 'NEK4': 17, 'NEK5': 17, 'NEK6': 17, 'NEK7': 17, 'NEK8': 17, 'NEK9': 17, 'NIK': 17, 'NIM1': 17, 'NLK': 17, 'NUAK1': 17, 'NUAK2': 17, 'OSR1': 17, 'P38A': 17, 'P38B': 17, 'P38D': 17, 'P38G': 17, 'P70S6K': 17, 'P70S6KB': 17, 'PAK1': 17, 'PAK2': 17, 'PAK3': 17, 'PAK4': 17, 'PAK5': 17, 'PAK6': 17, 'PASK': 17, 'PBK': 17, 'CDK16': 17, 'CDK17': 17, 'CDK18': 17, 'PDHK1': 16, 'PDHK4': 16, 'PDK1': 17, 'PERK': 17, 'CDK14': 17, 'PHKG1': 17, 'PHKG2': 17, 'PIM1': 17, 'PIM2': 17, 'PIM3': 17, 'PINK1': 17, 'PKACA': 17, 'PKACB': 17, 'PKACG': 17, 'PKCA': 17, 'PKCB': 17, 'PKCD': 17, 'PKCE': 17, 'PKCG': 17, 'PKCH': 17, 'PKCI': 17, 'PKCT': 17, 'PKCZ': 17, 'PRKD1': 17, 'PRKD2': 17, 'PRKD3': 17, 'PKG1': 17, 'PKG2': 17, 'PKN1': 17, 'PKN2': 17, 'PKN3': 17, 'PKR': 17, 'PLK1': 17, 'PLK2': 17, 'PLK3': 17, 'PLK4': 17, 'PRKX': 17, 'PRP4': 17, 'PRPK': 17, 'QIK': 17, 'QSK': 17, 'RAF1': 17, 'GRK1': 17, 'RIPK1': 17, 'RIPK2': 17, 'RIPK3': 17, 'ROCK1': 17, 'ROCK2': 17, 'P90RSK': 17, 'RSK2': 17, 'RSK3': 17, 'RSK4': 17, 'SBK': 17, 'MYLK4': 17, 'SGK1': 17, 'SGK3': 17, 'DSTYK': 17, 'SIK': 17, 'SKMLCK': 17, 'SLK': 17, 'SMG1': 17, 'SMMLCK': 17, 'SNRK': 17, 'SRPK1': 17, 'SRPK2': 17, 'SSTK': 17, 'STK33': 17, 'STLK3': 17, 'TAK1': 17, 'TAO1': 17, 'TAO2': 17, 'TAO3': 17, 'TBK1': 17, 'TGFBR1': 17, 'TGFBR2': 17, 'TLK1': 17, 'TLK2': 17, 'TNIK': 17, 'TSSK1': 17, 'TSSK2': 17, 'TTBK1': 17, 'TTBK2': 17, 'TTK': 17, 'ULK1': 17, 'ULK2': 17, 'VRK1': 17, 'VRK2': 17, 'WNK1': 17, 'WNK3': 17, 'WNK4': 17, 'YANK2': 17, 'YANK3': 17, 'YSK1': 17, 'ZAK': 17, 'EEF2K': 17, 'FAM20C': 17})
Multiply values, consider the dynamics of scale factor, which is PSPA random aa number.
get_pos_range (pssms_dict)
Get min and max position given a pssms_dict.
cut_seq_on_pssms (site_seq, pssms_dict)
Based on one pssm from pssms_dict, cut site seq if it is out of bound.
Let sequence be within the position range of reference PSSMs: -7 to +7.'PSVEPPLsQETFSDL'cut_seq_on_pssms_df (df, seq_col, pssms_dict)
Based on one pssm from pssms_dict, cut sequences in a df if it is out of bound.
Let sequence be within the position range of reference PSSMs: -7 to +7.0 ITGSRLLsMVPGPAR
1 VDDEKGDsNDDYDSA
2 YDSAGLLsDEDCMSV
3 IADHLFWsEETKSRF
4 KSRFTEYsMTssVMR
...
121327 EGGAGDRsEEEAsst
121328 DRsEEEAsstEKPtK
121329 RsEEEAsstEKPtKA
121330 sEEEAsstEKPtKAL
121331 AsstEKPtKALPRKS
Name: site_seq, Length: 119955, dtype: objectcalculate_log_odds (cut_seq, pssms_dict, site_type=None, bg_pssm=None, sort=True)
Calculate log odds based on cut sequence within the reference pssm range.
| Type | Default | Details | |
|---|---|---|---|
| cut_seq | site sequence to be scored | ||
| pssms_dict | key as kinase and value as flattened pssm | ||
| site_type | NoneType | None | |
| bg_pssm | NoneType | None | |
| sort | bool | True |
ATR 13.379067
ATM 11.133979
DNAPK 6.591411
CDK8 1.854333
TSSK1 1.621422
...
LIMK2 -20.450229
CDK3 -21.398831
SLK -23.802818
MRCKB -26.362947
PERK -27.007599
Length: 289, dtype: float64get_kinase_log_odds (site_seq, pssms_dict, **kwargs)
Calculate kinase score of a site sequence given pssms_dict and background pssm.
| Type | Details | |
|---|---|---|
| site_seq | site sequence to be scored | |
| pssms_dict | key as kinase and value as flattened pssm | |
| kwargs | VAR_KEYWORD |
Let sequence be within the position range of reference PSSMs: -7 to +7.ATR 13.379067
ATM 11.133979
DNAPK 6.591411
CDK8 1.854333
TSSK1 1.621422
...
LIMK2 -20.450229
CDK3 -21.398831
SLK -23.802818
MRCKB -26.362947
PERK -27.007599
Length: 289, dtype: float64check_seqs (seqs:pandas.core.series.Series)
Convert non-s/t/y to upper case & replace with underscore if the character is not in the allowed set
get_kinase_log_odds_df (df, seq_col, pssms_dict, parallel=True, sort=False, **kwargs)
Calculate kinase score of sequences in a df given pssms_dict and background pssm.
| Type | Default | Details | |
|---|---|---|---|
| df | |||
| seq_col | site sequence to be scored | ||
| pssms_dict | key as kinase and value as flattened pssm | ||
| parallel | bool | True | use parallel processing if True |
| sort | bool | False | |
| kwargs | VAR_KEYWORD |
Let sequence be within the position range of reference PSSMs: -7 to +7.
0%| | 0/10 [00:00<?, ?it/s]
100%|██████████| 10/10 [00:00<00:00, 81.17it/s][A| SRC | EPHA3 | FES | NTRK3 | ALK | EPHA8 | ABL1 | FLT3 | EPHB2 | FYN | ... | MEK5 | PKN2 | MAP2K7 | MRCKB | HIPK3 | CDK8 | BUB1 | MEKK3 | MAP2K3 | GRK1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -6.834939 | -6.226365 | -7.695221 | -5.778931 | -6.748094 | -7.305816 | -4.949184 | -6.795048 | -6.780491 | -7.430980 | ... | -13.751513 | -6.700875 | 0.664614 | -11.965351 | -8.269410 | -15.987455 | -3.024525 | -12.537066 | 2.067269 | -34.978009 |
| 1 | -2.787712 | -2.184116 | -2.030497 | -3.016946 | -2.795443 | -1.633924 | -3.606747 | -3.197927 | -2.535484 | -1.439454 | ... | -9.929361 | -13.842463 | -10.501498 | -8.838672 | -28.157528 | -7.236805 | -12.967786 | -10.855866 | -9.532418 | -20.914510 |
| 2 | -4.400023 | -3.880026 | -3.600108 | -3.433173 | -4.822332 | -5.214294 | -5.780404 | -6.425483 | -4.559895 | -4.087982 | ... | -34.949360 | -20.840285 | -27.924034 | -39.232552 | -13.082519 | -21.271829 | -16.060199 | -15.896029 | -26.803760 | -10.950010 |
| 3 | -1.858100 | -2.077535 | -3.137042 | -1.826046 | -2.842460 | -3.105884 | -2.904125 | -2.888445 | -2.601997 | -2.483656 | ... | -7.028202 | 6.532928 | -0.672702 | -28.807424 | -44.272850 | -11.192035 | -10.501577 | -21.918909 | -9.375008 | -5.354777 |
| 4 | -7.846949 | -9.211063 | -10.707947 | -10.049246 | -11.106238 | -10.004203 | -8.335588 | -17.250129 | -11.380093 | -10.607860 | ... | -45.751185 | -33.215923 | -7.332241 | -30.562655 | -44.583823 | -36.421097 | -24.849370 | -48.512109 | -31.798307 | -44.706866 |
| 5 | -5.573933 | -6.375702 | -6.180108 | -5.660873 | -6.094550 | -6.110192 | -5.577982 | -6.640383 | -6.049525 | -5.108223 | ... | -24.883888 | -25.395896 | -10.664401 | -21.363969 | -23.513679 | -24.974486 | -9.292972 | -23.527402 | -17.512193 | -40.701488 |
| 6 | -8.135098 | -6.596200 | -8.303372 | -7.374844 | -6.232523 | -7.147063 | -6.242102 | -5.788387 | -7.126718 | -7.285538 | ... | -12.880634 | -25.752945 | -20.574576 | -39.697786 | -33.637365 | -31.118501 | -36.537491 | -24.841079 | -17.103741 | -32.904123 |
| 7 | -8.618104 | -11.024002 | -10.093951 | -9.237905 | -9.946672 | -11.984708 | -9.190825 | -9.362905 | -9.830988 | -9.136566 | ... | -16.486060 | -12.437818 | 1.196805 | -17.744377 | 3.511220 | -22.600149 | -4.465207 | -16.292567 | 0.798599 | -15.486682 |
| 8 | -6.127544 | -7.267991 | -7.159930 | -5.897895 | -6.461399 | -7.633443 | -5.801562 | -7.011265 | -7.294982 | -6.769097 | ... | -3.700738 | 1.147968 | -0.781214 | -1.852455 | -3.598562 | -4.785045 | -3.083286 | -1.962302 | -2.607863 | -17.613164 |
| 9 | -2.679757 | -3.688841 | -3.617050 | -2.983916 | -3.287914 | -3.672047 | -2.311731 | -2.951490 | -2.286616 | -2.913383 | ... | -35.733420 | -6.279436 | -16.687181 | -18.868566 | -4.874849 | -38.264279 | -10.590697 | -15.189655 | -10.709765 | -39.215480 |
10 rows × 289 columns
sumup (values, kinase=None)
Sum up the possibilities of the amino acids at each position in a phosphorylation site sequence
| Type | Default | Details | |
|---|---|---|---|
| values | list of values, possibilities of amino acids at certain positions | ||
| kinase | NoneType | None |
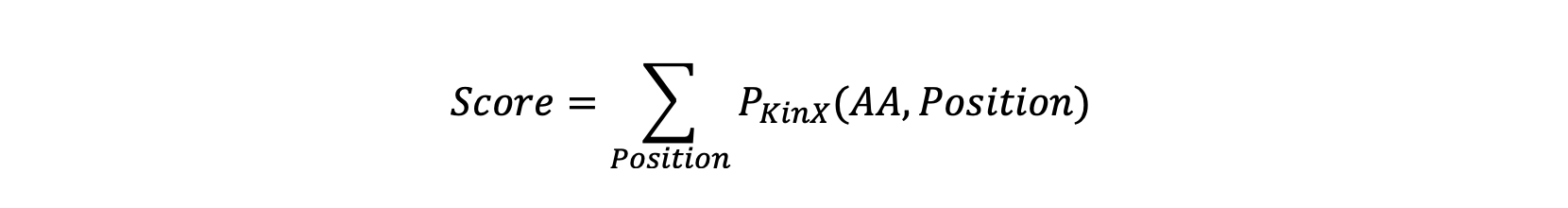
predict_kinase (input_string:str, ref:pandas.core.frame.DataFrame, func:Callable, to_lower:bool=False, to_upper:bool=False, verbose=True)
Predict kinase given a phosphorylation site sequence
| Type | Default | Details | |
|---|---|---|---|
| input_string | str | site sequence | |
| ref | DataFrame | reference dataframe for scoring | |
| func | Callable | function to calculate score | |
| to_lower | bool | False | convert capital STY to lower case |
| to_upper | bool | False | convert all letter to uppercase |
| verbose | bool | True |
considering string: ['-5V', '-4E', '-3P', '-2P', '-1L', '0s', '1Q', '2E', '3T', '4F']kinase
ATM 0.167
SMG1 -0.060
DNAPK -0.714
FAM20C -1.216
ATR -1.321
...
PKCI -11.319
NEK3 -11.455
CK1A -11.686
CK1G3 -13.182
CK1G2 -13.421
Length: 303, dtype: float64Here we provide different PSSM settings from either PSPA data or kinase-substrate dataset for kinase prediction:
Params (name=None)
considering string: ['-5V', '-4E', '-3P', '-2P', '-1L', '0s', '1Q', '2E', '3T', '4F', '5S']
kinase
ATM 5.037
SMG1 4.385
DNAPK 3.818
ATR 3.507
FAM20C 3.170
dtype: float64
considering string: ['-7P', '-6S', '-5V', '-4E', '-3P', '-2P', '-1L', '0s', '1Q', '2E', '3T', '4F', '5S', '6D', '7L']
kinase
ATR 3.064
ATM 2.909
DNAPK 2.270
CK2A1 1.873
TSSK1 1.856
dtype: float64
considering string: ['-7P', '-6S', '-5V', '-4E', '-3P', '-2P', '-1L', '0S', '1Q', '2E', '3T', '4F', '5S', '6D', '7L']
kinase
ATR 3.229
ATM 3.038
DNAPK 2.479
CK2A1 2.006
CDK8 1.999
dtype: float64predict_kinase_df (df, seq_col, ref, func, to_lower=False, to_upper=False)
get_pct (site, ref, func, pct_ref)
Replicate the precentile results from The Kinase Library.
out = get_pct('PSVEPPLyQETFSDL',**Params('PSPA_y'), pct_ref=y_pct)
out.sort_values('percentile',ascending=False)considering string: ['-5V', '-4E', '-3P', '-2P', '-1L', '0Y', '1Q', '2E', '3T', '4F', '5S']| log2(score) | percentile | |
|---|---|---|
| ABL2 | 3.137 | 96.568694 |
| BMX | 2.816 | 96.117567 |
| BTK | 1.956 | 95.693780 |
| CSK | 2.303 | 95.174299 |
| MERTK | 2.509 | 93.588517 |
| ... | ... | ... |
| FLT1 | -1.919 | 25.358852 |
| PINK1_TYR | -1.227 | 21.927546 |
| MUSK | -3.031 | 21.298701 |
| TNNI3K_TYR | -3.549 | 11.004785 |
| PKMYT1_TYR | -1.739 | 4.798360 |
93 rows × 2 columns
considering string: ['-5V', '-4E', '-3P', '-2P', '-1L', '0S', '1Q', '2E', '3T', '4F']| log2(score) | percentile | |
|---|---|---|
| ATM | 5.037 | 99.822351 |
| SMG1 | 4.385 | 99.831819 |
| DNAPK | 3.818 | 99.205315 |
| ATR | 3.507 | 99.680344 |
| FAM20C | 3.170 | 95.370556 |
| ... | ... | ... |
| PKN1 | -7.275 | 14.070436 |
| P70S6K | -7.295 | 4.089816 |
| AKT3 | -7.375 | 11.432995 |
| PKCI | -7.742 | 8.129511 |
| NEK3 | -8.254 | 4.637240 |
303 rows × 2 columns
get_pct_df (score_df, pct_ref)
Replicate the precentile results from The Kinase Library.
| Details | |
|---|---|
| score_df | output from predict_kinase_df |
| pct_ref | a reference df for percentile calculation |