from katlas.core import *
import pandas as pdBackground frequencies
df = Data.get_ks_unique()df.head()| sub_site | num_kin | bin | sub_genes | site_seq | source_combine | acceptor | O00141_SGK1 | O00238_BMPR1B | O00311_CDC7 | ... | Q9Y2K2_SIK3 | Q9Y2U5_MAP3K2 | Q9Y3S1_WNK2 | Q9Y463_DYRK1B | Q9Y4K4_MAP4K5 | Q9Y572_RIPK3 | Q9Y5S2_CDC42BPB | Q9Y6E0_STK24 | Q9Y6M4_CSNK1G3 | Q9Y6R4_MAP3K4 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | A0A2R8Y4L2_S158 | 1 | 1 | HNRNPA1L3 HNRNPA1P48 | TDRGSGKKRGFAFVTFDDHDsVDKIVIQKYHTVNGHNCEVR | Sugiyama | S | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | A0A2R8Y4L2_S22 | 3 | 2~10 | HNRNPA1L3 HNRNPA1P48 | SKSEsPKEPEQLRKLFIGGLsFEtTDESLRSHFEQWGTLTD | Sugiyama | S | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2 | A0A2R8Y4L2_S6 | 3 | 2~10 | HNRNPA1L3 HNRNPA1P48 | _______________MSKSEsPKEPEQLRKLFIGGLsFEtT | Sugiyama | S | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | A0A2R8Y4L2_S95 | 65 | 11~100 | HNRNPA1L3 HNRNPA1P48 | RPHKVDGRVVEPKRAVSREDsQRPDAHLTVKKIFVGGIKED | Sugiyama | S | 0 | 1 | 0 | ... | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 4 | A0A2R8Y4L2_T25 | 3 | 2~10 | HNRNPA1L3 HNRNPA1P48 | EsPKEPEQLRKLFIGGLsFEtTDESLRSHFEQWGTLTDCVV | Sugiyama | T | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
5 rows × 462 columns
PSSMs - ks dataset
def get_bg_dict(df, acceptor, seq_col='site_seq'):
site = df[df.acceptor.isin(list(acceptor))].copy()
site_pssm = get_prob(site, seq_col)
return flatten_pssm(site_pssm, True)acceptors = ['S','T','Y','ST','STY']index_names = 'ks_' + pd.Series(acceptors)bg_pssms = pd.DataFrame([get_bg_dict(df,acceptor) for acceptor in acceptors],index=index_names)bg_pssms| -20P | -20G | -20A | -20C | -20S | -20T | -20V | -20I | -20L | -20M | ... | 20H | 20K | 20R | 20Q | 20N | 20D | 20E | 20s | 20t | 20y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ks_S | 0.074296 | 0.068105 | 0.075925 | 0.014664 | 0.053246 | 0.038517 | 0.051290 | 0.040342 | 0.078989 | 0.020269 | ... | 0.021294 | 0.070330 | 0.059313 | 0.044200 | 0.036273 | 0.054880 | 0.077316 | 0.043461 | 0.014980 | 0.006449 |
| ks_T | 0.061634 | 0.065896 | 0.074266 | 0.016436 | 0.043829 | 0.034394 | 0.059656 | 0.040177 | 0.089484 | 0.024806 | ... | 0.022394 | 0.075067 | 0.060558 | 0.045261 | 0.038007 | 0.058666 | 0.076644 | 0.035483 | 0.019871 | 0.008989 |
| ks_Y | 0.056181 | 0.072421 | 0.070666 | 0.015947 | 0.044477 | 0.038771 | 0.055889 | 0.045940 | 0.086320 | 0.024287 | ... | 0.019500 | 0.071146 | 0.057892 | 0.042505 | 0.037629 | 0.056216 | 0.083181 | 0.023157 | 0.012492 | 0.017367 |
| ks_ST | 0.070500 | 0.067442 | 0.075428 | 0.015195 | 0.050422 | 0.037280 | 0.053799 | 0.040292 | 0.082136 | 0.021629 | ... | 0.021622 | 0.071745 | 0.059685 | 0.044517 | 0.036791 | 0.056011 | 0.077115 | 0.041078 | 0.016441 | 0.007207 |
| ks_STY | 0.067096 | 0.068626 | 0.074296 | 0.015374 | 0.049009 | 0.037635 | 0.054296 | 0.041635 | 0.083130 | 0.022261 | ... | 0.021121 | 0.071603 | 0.059262 | 0.044041 | 0.036989 | 0.056059 | 0.078548 | 0.036845 | 0.015508 | 0.009607 |
5 rows × 943 columns
index_names_upper = 'ks_' + pd.Series(acceptors) + '_upper'df['site_seq_upper'] = df['site_seq'].str.upper()bg_pssms_upper = pd.DataFrame([get_bg_dict(df,acceptor,seq_col='site_seq_upper') for acceptor in acceptors],index=index_names_upper)bg_pssms = pd.concat([bg_pssms,bg_pssms_upper])bg_pssms| -20P | -20G | -20A | -20C | -20S | -20T | -20V | -20I | -20L | -20M | ... | 20H | 20K | 20R | 20Q | 20N | 20D | 20E | 20s | 20t | 20y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ks_S | 0.074296 | 0.068105 | 0.075925 | 0.014664 | 0.053246 | 0.038517 | 0.051290 | 0.040342 | 0.078989 | 0.020269 | ... | 0.021294 | 0.070330 | 0.059313 | 0.044200 | 0.036273 | 0.054880 | 0.077316 | 0.043461 | 0.014980 | 0.006449 |
| ks_T | 0.061634 | 0.065896 | 0.074266 | 0.016436 | 0.043829 | 0.034394 | 0.059656 | 0.040177 | 0.089484 | 0.024806 | ... | 0.022394 | 0.075067 | 0.060558 | 0.045261 | 0.038007 | 0.058666 | 0.076644 | 0.035483 | 0.019871 | 0.008989 |
| ks_Y | 0.056181 | 0.072421 | 0.070666 | 0.015947 | 0.044477 | 0.038771 | 0.055889 | 0.045940 | 0.086320 | 0.024287 | ... | 0.019500 | 0.071146 | 0.057892 | 0.042505 | 0.037629 | 0.056216 | 0.083181 | 0.023157 | 0.012492 | 0.017367 |
| ks_ST | 0.070500 | 0.067442 | 0.075428 | 0.015195 | 0.050422 | 0.037280 | 0.053799 | 0.040292 | 0.082136 | 0.021629 | ... | 0.021622 | 0.071745 | 0.059685 | 0.044517 | 0.036791 | 0.056011 | 0.077115 | 0.041078 | 0.016441 | 0.007207 |
| ks_STY | 0.067096 | 0.068626 | 0.074296 | 0.015374 | 0.049009 | 0.037635 | 0.054296 | 0.041635 | 0.083130 | 0.022261 | ... | 0.021121 | 0.071603 | 0.059262 | 0.044041 | 0.036989 | 0.056059 | 0.078548 | 0.036845 | 0.015508 | 0.009607 |
| ks_S_upper | 0.074296 | 0.068105 | 0.075925 | 0.014664 | 0.094891 | 0.054028 | 0.051290 | 0.040342 | 0.078989 | 0.020269 | ... | 0.021294 | 0.070330 | 0.059313 | 0.044200 | 0.036273 | 0.054880 | 0.077316 | 0.000000 | 0.000000 | 0.000000 |
| ks_T_upper | 0.061634 | 0.065896 | 0.074266 | 0.016436 | 0.072744 | 0.052960 | 0.059656 | 0.040177 | 0.089484 | 0.024806 | ... | 0.022394 | 0.075067 | 0.060558 | 0.045261 | 0.038007 | 0.058666 | 0.076644 | 0.000000 | 0.000000 | 0.000000 |
| ks_Y_upper | 0.056181 | 0.072421 | 0.070666 | 0.015947 | 0.067008 | 0.050475 | 0.055889 | 0.045940 | 0.086320 | 0.024287 | ... | 0.019500 | 0.071146 | 0.057892 | 0.042505 | 0.037629 | 0.056216 | 0.083181 | 0.000000 | 0.000000 | 0.000000 |
| ks_ST_upper | 0.070500 | 0.067442 | 0.075428 | 0.015195 | 0.088250 | 0.053708 | 0.053799 | 0.040292 | 0.082136 | 0.021629 | ... | 0.021622 | 0.071745 | 0.059685 | 0.044517 | 0.036791 | 0.056011 | 0.077115 | 0.000000 | 0.000000 | 0.000000 |
| ks_STY_upper | 0.067096 | 0.068626 | 0.074296 | 0.015374 | 0.083200 | 0.052939 | 0.054296 | 0.041635 | 0.083130 | 0.022261 | ... | 0.021121 | 0.071603 | 0.059262 | 0.044041 | 0.036989 | 0.056059 | 0.078548 | 0.000000 | 0.000000 | 0.000000 |
10 rows × 943 columns
def get_site_cnt(df, acceptor, seq_col='site_seq'):
site = df[df.acceptor.isin(list(acceptor))].copy()
return len(site)cnt = {name:get_site_cnt(df,acceptor) for acceptor,name in zip(acceptors,index_names)}plot_logos(bg_pssms,cnt,prefix='')
save_pdf('fig/background_ks.pdf')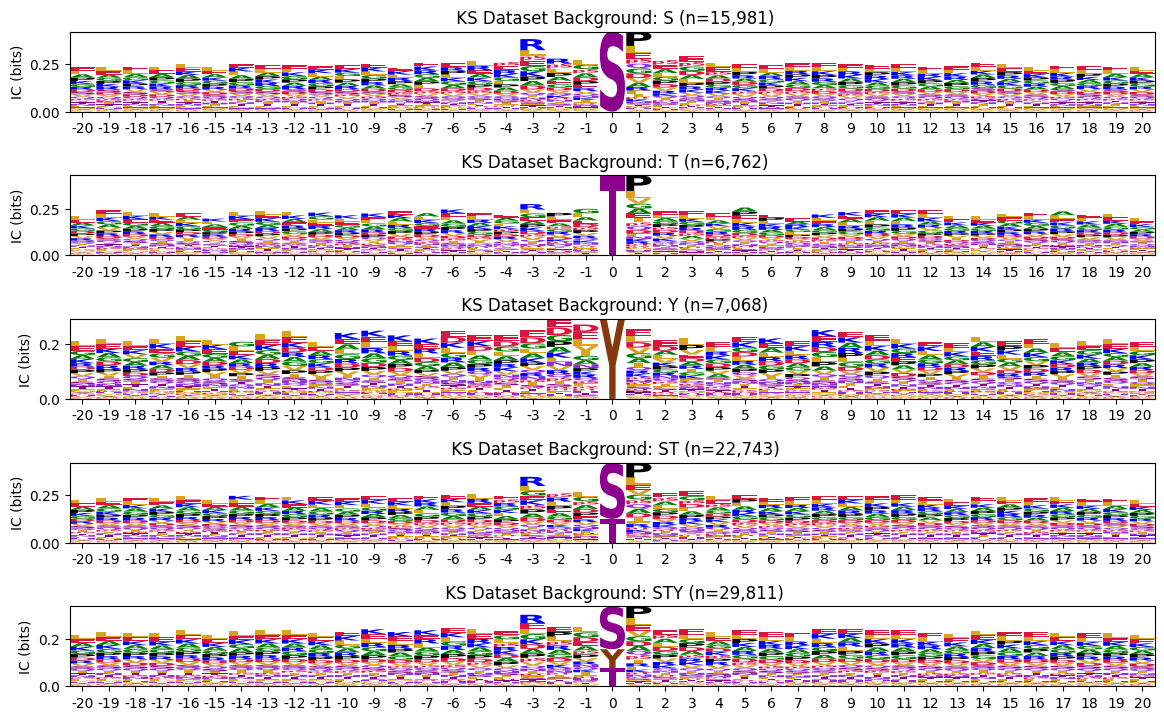
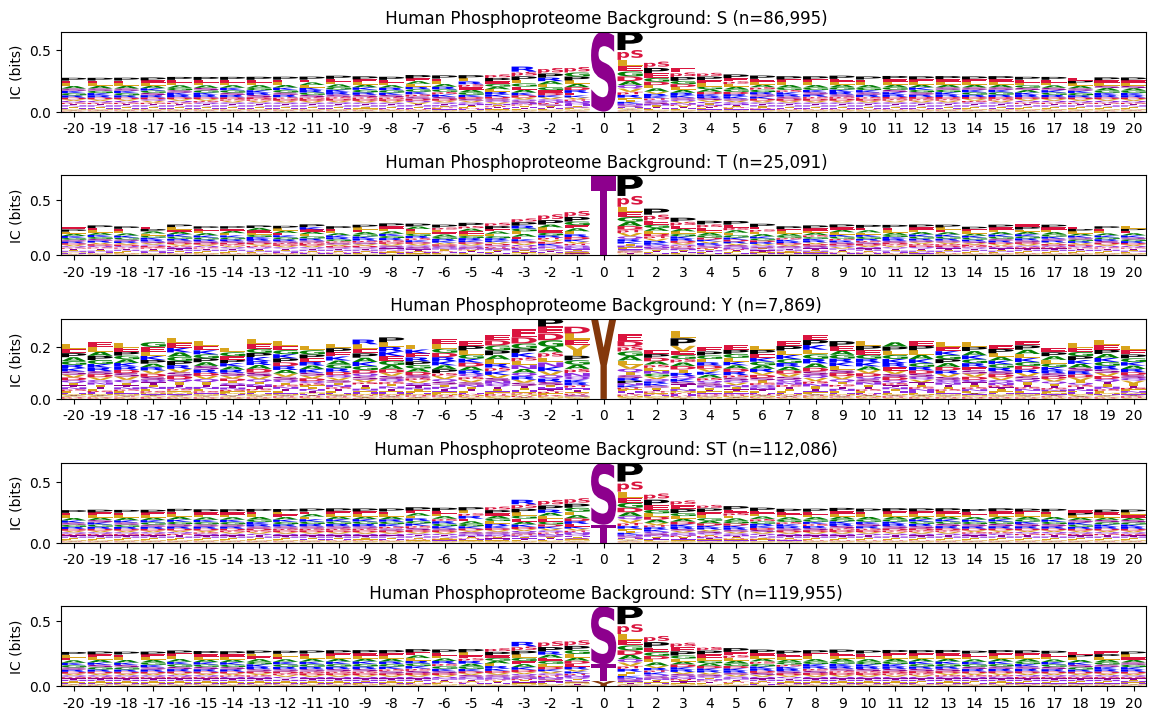
PSSMs - human phosphoproteome
human = Data.get_human_site()human['acceptor']=human.site.str[0]human.head(1)| substrate_uniprot | substrate_genes | site | source | AM_pathogenicity | substrate_sequence | substrate_species | sub_site | substrate_phosphoseq | position | site_seq | acceptor | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | A0A024R4G9 | C19orf48 MGC13170 hCG_2008493 | S20 | psp | NaN | MTVLEAVLEIQAITGSRLLSMVPGPARPPGSCWDPTQCTRTWLLSH... | Homo sapiens (Human) | A0A024R4G9_S20 | MTVLEAVLEIQAITGSRLLsMVPGPARPPGSCWDPTQCTRTWLLSH... | 20 | _MTVLEAVLEIQAITGSRLLsMVPGPARPPGSCWDPTQCTR | S |
index_names2 = 'human_' + pd.Series(acceptors)bg_pssms_human = pd.DataFrame([get_bg_dict(human,acceptor) for acceptor in acceptors],index=index_names2)bg_pssms_human| -20P | -20G | -20A | -20C | -20S | -20T | -20V | -20I | -20L | -20M | ... | 20H | 20K | 20R | 20Q | 20N | 20D | 20E | 20s | 20t | 20y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| human_S | 0.083640 | 0.067324 | 0.072426 | 0.012983 | 0.068431 | 0.040755 | 0.048601 | 0.032621 | 0.080464 | 0.018531 | ... | 0.022398 | 0.066432 | 0.064524 | 0.047369 | 0.033162 | 0.051790 | 0.079818 | 0.044602 | 0.013168 | 0.003673 |
| human_T | 0.079864 | 0.064356 | 0.074764 | 0.011942 | 0.062697 | 0.040927 | 0.052994 | 0.033463 | 0.078537 | 0.021811 | ... | 0.020073 | 0.070087 | 0.062644 | 0.048049 | 0.032200 | 0.051813 | 0.078660 | 0.043658 | 0.017020 | 0.004851 |
| human_Y | 0.073897 | 0.067143 | 0.073103 | 0.016024 | 0.058403 | 0.039465 | 0.051781 | 0.041451 | 0.087406 | 0.020262 | ... | 0.021772 | 0.065723 | 0.059600 | 0.049939 | 0.031297 | 0.055382 | 0.078106 | 0.034699 | 0.016193 | 0.016601 |
| human_ST | 0.082791 | 0.066656 | 0.072951 | 0.012749 | 0.067141 | 0.040794 | 0.049589 | 0.032810 | 0.080030 | 0.019268 | ... | 0.021877 | 0.067252 | 0.064102 | 0.047521 | 0.032946 | 0.051795 | 0.079558 | 0.044391 | 0.014031 | 0.003937 |
| human_STY | 0.082206 | 0.066688 | 0.072961 | 0.012965 | 0.066566 | 0.040706 | 0.049733 | 0.033379 | 0.080515 | 0.019334 | ... | 0.021870 | 0.067153 | 0.063812 | 0.047677 | 0.032840 | 0.052027 | 0.079465 | 0.043766 | 0.014171 | 0.004753 |
5 rows × 943 columns
index_names2_upper = 'human_' + pd.Series(acceptors) +'_upper'human['site_seq_upper'] = human['site_seq'].str.upper()bg_pssms_human_upper = pd.DataFrame([get_bg_dict(human,acceptor,seq_col='site_seq_upper') for acceptor in acceptors],index=index_names2_upper)bg_pssms_human_upper| -20P | -20G | -20A | -20C | -20S | -20T | -20V | -20I | -20L | -20M | ... | 20H | 20K | 20R | 20Q | 20N | 20D | 20E | 20s | 20t | 20y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| human_S_upper | 0.083640 | 0.067324 | 0.072426 | 0.012983 | 0.112772 | 0.053678 | 0.048601 | 0.032621 | 0.080464 | 0.018531 | ... | 0.022398 | 0.066432 | 0.064524 | 0.047369 | 0.033162 | 0.051790 | 0.079818 | 0.0 | 0.0 | 0.0 |
| human_T_upper | 0.079864 | 0.064356 | 0.074764 | 0.011942 | 0.106734 | 0.058260 | 0.052994 | 0.033463 | 0.078537 | 0.021811 | ... | 0.020073 | 0.070087 | 0.062644 | 0.048049 | 0.032200 | 0.051813 | 0.078660 | 0.0 | 0.0 | 0.0 |
| human_Y_upper | 0.073897 | 0.067143 | 0.073103 | 0.016024 | 0.093630 | 0.053900 | 0.051781 | 0.041451 | 0.087406 | 0.020262 | ... | 0.021772 | 0.065723 | 0.059600 | 0.049939 | 0.031297 | 0.055382 | 0.078106 | 0.0 | 0.0 | 0.0 |
| human_ST_upper | 0.082791 | 0.066656 | 0.072951 | 0.012749 | 0.111414 | 0.054709 | 0.049589 | 0.032810 | 0.080030 | 0.019268 | ... | 0.021877 | 0.067252 | 0.064102 | 0.047521 | 0.032946 | 0.051795 | 0.079558 | 0.0 | 0.0 | 0.0 |
| human_STY_upper | 0.082206 | 0.066688 | 0.072961 | 0.012965 | 0.110244 | 0.054656 | 0.049733 | 0.033379 | 0.080515 | 0.019334 | ... | 0.021870 | 0.067153 | 0.063812 | 0.047677 | 0.032840 | 0.052027 | 0.079465 | 0.0 | 0.0 | 0.0 |
5 rows × 943 columns
bg_pssms_human = pd.concat([bg_pssms_human,bg_pssms_human_upper])bg_pssms_human| -20P | -20G | -20A | -20C | -20S | -20T | -20V | -20I | -20L | -20M | ... | 20H | 20K | 20R | 20Q | 20N | 20D | 20E | 20s | 20t | 20y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| human_S | 0.083640 | 0.067324 | 0.072426 | 0.012983 | 0.068431 | 0.040755 | 0.048601 | 0.032621 | 0.080464 | 0.018531 | ... | 0.022398 | 0.066432 | 0.064524 | 0.047369 | 0.033162 | 0.051790 | 0.079818 | 0.044602 | 0.013168 | 0.003673 |
| human_T | 0.079864 | 0.064356 | 0.074764 | 0.011942 | 0.062697 | 0.040927 | 0.052994 | 0.033463 | 0.078537 | 0.021811 | ... | 0.020073 | 0.070087 | 0.062644 | 0.048049 | 0.032200 | 0.051813 | 0.078660 | 0.043658 | 0.017020 | 0.004851 |
| human_Y | 0.073897 | 0.067143 | 0.073103 | 0.016024 | 0.058403 | 0.039465 | 0.051781 | 0.041451 | 0.087406 | 0.020262 | ... | 0.021772 | 0.065723 | 0.059600 | 0.049939 | 0.031297 | 0.055382 | 0.078106 | 0.034699 | 0.016193 | 0.016601 |
| human_ST | 0.082791 | 0.066656 | 0.072951 | 0.012749 | 0.067141 | 0.040794 | 0.049589 | 0.032810 | 0.080030 | 0.019268 | ... | 0.021877 | 0.067252 | 0.064102 | 0.047521 | 0.032946 | 0.051795 | 0.079558 | 0.044391 | 0.014031 | 0.003937 |
| human_STY | 0.082206 | 0.066688 | 0.072961 | 0.012965 | 0.066566 | 0.040706 | 0.049733 | 0.033379 | 0.080515 | 0.019334 | ... | 0.021870 | 0.067153 | 0.063812 | 0.047677 | 0.032840 | 0.052027 | 0.079465 | 0.043766 | 0.014171 | 0.004753 |
| human_S_upper | 0.083640 | 0.067324 | 0.072426 | 0.012983 | 0.112772 | 0.053678 | 0.048601 | 0.032621 | 0.080464 | 0.018531 | ... | 0.022398 | 0.066432 | 0.064524 | 0.047369 | 0.033162 | 0.051790 | 0.079818 | 0.000000 | 0.000000 | 0.000000 |
| human_T_upper | 0.079864 | 0.064356 | 0.074764 | 0.011942 | 0.106734 | 0.058260 | 0.052994 | 0.033463 | 0.078537 | 0.021811 | ... | 0.020073 | 0.070087 | 0.062644 | 0.048049 | 0.032200 | 0.051813 | 0.078660 | 0.000000 | 0.000000 | 0.000000 |
| human_Y_upper | 0.073897 | 0.067143 | 0.073103 | 0.016024 | 0.093630 | 0.053900 | 0.051781 | 0.041451 | 0.087406 | 0.020262 | ... | 0.021772 | 0.065723 | 0.059600 | 0.049939 | 0.031297 | 0.055382 | 0.078106 | 0.000000 | 0.000000 | 0.000000 |
| human_ST_upper | 0.082791 | 0.066656 | 0.072951 | 0.012749 | 0.111414 | 0.054709 | 0.049589 | 0.032810 | 0.080030 | 0.019268 | ... | 0.021877 | 0.067252 | 0.064102 | 0.047521 | 0.032946 | 0.051795 | 0.079558 | 0.000000 | 0.000000 | 0.000000 |
| human_STY_upper | 0.082206 | 0.066688 | 0.072961 | 0.012965 | 0.110244 | 0.054656 | 0.049733 | 0.033379 | 0.080515 | 0.019334 | ... | 0.021870 | 0.067153 | 0.063812 | 0.047677 | 0.032840 | 0.052027 | 0.079465 | 0.000000 | 0.000000 | 0.000000 |
10 rows × 943 columns
all_pssms = pd.concat([bg_pssms,bg_pssms_human])all_pssms| -20P | -20G | -20A | -20C | -20S | -20T | -20V | -20I | -20L | -20M | ... | 20H | 20K | 20R | 20Q | 20N | 20D | 20E | 20s | 20t | 20y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ks_S | 0.074296 | 0.068105 | 0.075925 | 0.014664 | 0.053246 | 0.038517 | 0.051290 | 0.040342 | 0.078989 | 0.020269 | ... | 0.021294 | 0.070330 | 0.059313 | 0.044200 | 0.036273 | 0.054880 | 0.077316 | 0.043461 | 0.014980 | 0.006449 |
| ks_T | 0.061634 | 0.065896 | 0.074266 | 0.016436 | 0.043829 | 0.034394 | 0.059656 | 0.040177 | 0.089484 | 0.024806 | ... | 0.022394 | 0.075067 | 0.060558 | 0.045261 | 0.038007 | 0.058666 | 0.076644 | 0.035483 | 0.019871 | 0.008989 |
| ks_Y | 0.056181 | 0.072421 | 0.070666 | 0.015947 | 0.044477 | 0.038771 | 0.055889 | 0.045940 | 0.086320 | 0.024287 | ... | 0.019500 | 0.071146 | 0.057892 | 0.042505 | 0.037629 | 0.056216 | 0.083181 | 0.023157 | 0.012492 | 0.017367 |
| ks_ST | 0.070500 | 0.067442 | 0.075428 | 0.015195 | 0.050422 | 0.037280 | 0.053799 | 0.040292 | 0.082136 | 0.021629 | ... | 0.021622 | 0.071745 | 0.059685 | 0.044517 | 0.036791 | 0.056011 | 0.077115 | 0.041078 | 0.016441 | 0.007207 |
| ks_STY | 0.067096 | 0.068626 | 0.074296 | 0.015374 | 0.049009 | 0.037635 | 0.054296 | 0.041635 | 0.083130 | 0.022261 | ... | 0.021121 | 0.071603 | 0.059262 | 0.044041 | 0.036989 | 0.056059 | 0.078548 | 0.036845 | 0.015508 | 0.009607 |
| ks_S_upper | 0.074296 | 0.068105 | 0.075925 | 0.014664 | 0.094891 | 0.054028 | 0.051290 | 0.040342 | 0.078989 | 0.020269 | ... | 0.021294 | 0.070330 | 0.059313 | 0.044200 | 0.036273 | 0.054880 | 0.077316 | 0.000000 | 0.000000 | 0.000000 |
| ks_T_upper | 0.061634 | 0.065896 | 0.074266 | 0.016436 | 0.072744 | 0.052960 | 0.059656 | 0.040177 | 0.089484 | 0.024806 | ... | 0.022394 | 0.075067 | 0.060558 | 0.045261 | 0.038007 | 0.058666 | 0.076644 | 0.000000 | 0.000000 | 0.000000 |
| ks_Y_upper | 0.056181 | 0.072421 | 0.070666 | 0.015947 | 0.067008 | 0.050475 | 0.055889 | 0.045940 | 0.086320 | 0.024287 | ... | 0.019500 | 0.071146 | 0.057892 | 0.042505 | 0.037629 | 0.056216 | 0.083181 | 0.000000 | 0.000000 | 0.000000 |
| ks_ST_upper | 0.070500 | 0.067442 | 0.075428 | 0.015195 | 0.088250 | 0.053708 | 0.053799 | 0.040292 | 0.082136 | 0.021629 | ... | 0.021622 | 0.071745 | 0.059685 | 0.044517 | 0.036791 | 0.056011 | 0.077115 | 0.000000 | 0.000000 | 0.000000 |
| ks_STY_upper | 0.067096 | 0.068626 | 0.074296 | 0.015374 | 0.083200 | 0.052939 | 0.054296 | 0.041635 | 0.083130 | 0.022261 | ... | 0.021121 | 0.071603 | 0.059262 | 0.044041 | 0.036989 | 0.056059 | 0.078548 | 0.000000 | 0.000000 | 0.000000 |
| human_S | 0.083640 | 0.067324 | 0.072426 | 0.012983 | 0.068431 | 0.040755 | 0.048601 | 0.032621 | 0.080464 | 0.018531 | ... | 0.022398 | 0.066432 | 0.064524 | 0.047369 | 0.033162 | 0.051790 | 0.079818 | 0.044602 | 0.013168 | 0.003673 |
| human_T | 0.079864 | 0.064356 | 0.074764 | 0.011942 | 0.062697 | 0.040927 | 0.052994 | 0.033463 | 0.078537 | 0.021811 | ... | 0.020073 | 0.070087 | 0.062644 | 0.048049 | 0.032200 | 0.051813 | 0.078660 | 0.043658 | 0.017020 | 0.004851 |
| human_Y | 0.073897 | 0.067143 | 0.073103 | 0.016024 | 0.058403 | 0.039465 | 0.051781 | 0.041451 | 0.087406 | 0.020262 | ... | 0.021772 | 0.065723 | 0.059600 | 0.049939 | 0.031297 | 0.055382 | 0.078106 | 0.034699 | 0.016193 | 0.016601 |
| human_ST | 0.082791 | 0.066656 | 0.072951 | 0.012749 | 0.067141 | 0.040794 | 0.049589 | 0.032810 | 0.080030 | 0.019268 | ... | 0.021877 | 0.067252 | 0.064102 | 0.047521 | 0.032946 | 0.051795 | 0.079558 | 0.044391 | 0.014031 | 0.003937 |
| human_STY | 0.082206 | 0.066688 | 0.072961 | 0.012965 | 0.066566 | 0.040706 | 0.049733 | 0.033379 | 0.080515 | 0.019334 | ... | 0.021870 | 0.067153 | 0.063812 | 0.047677 | 0.032840 | 0.052027 | 0.079465 | 0.043766 | 0.014171 | 0.004753 |
| human_S_upper | 0.083640 | 0.067324 | 0.072426 | 0.012983 | 0.112772 | 0.053678 | 0.048601 | 0.032621 | 0.080464 | 0.018531 | ... | 0.022398 | 0.066432 | 0.064524 | 0.047369 | 0.033162 | 0.051790 | 0.079818 | 0.000000 | 0.000000 | 0.000000 |
| human_T_upper | 0.079864 | 0.064356 | 0.074764 | 0.011942 | 0.106734 | 0.058260 | 0.052994 | 0.033463 | 0.078537 | 0.021811 | ... | 0.020073 | 0.070087 | 0.062644 | 0.048049 | 0.032200 | 0.051813 | 0.078660 | 0.000000 | 0.000000 | 0.000000 |
| human_Y_upper | 0.073897 | 0.067143 | 0.073103 | 0.016024 | 0.093630 | 0.053900 | 0.051781 | 0.041451 | 0.087406 | 0.020262 | ... | 0.021772 | 0.065723 | 0.059600 | 0.049939 | 0.031297 | 0.055382 | 0.078106 | 0.000000 | 0.000000 | 0.000000 |
| human_ST_upper | 0.082791 | 0.066656 | 0.072951 | 0.012749 | 0.111414 | 0.054709 | 0.049589 | 0.032810 | 0.080030 | 0.019268 | ... | 0.021877 | 0.067252 | 0.064102 | 0.047521 | 0.032946 | 0.051795 | 0.079558 | 0.000000 | 0.000000 | 0.000000 |
| human_STY_upper | 0.082206 | 0.066688 | 0.072961 | 0.012965 | 0.110244 | 0.054656 | 0.049733 | 0.033379 | 0.080515 | 0.019334 | ... | 0.021870 | 0.067153 | 0.063812 | 0.047677 | 0.032840 | 0.052027 | 0.079465 | 0.000000 | 0.000000 | 0.000000 |
20 rows × 943 columns
# all_pssms.to_parquet('out/ks_background.parquet')Accessible through Data.get_ks_background
cnt_human = {name:get_site_cnt(human,acceptor) for acceptor,name in zip(acceptors,index_names2)}cnt.update(cnt_human)cnt{'KS Dataset Background: S': 15981,
'KS Dataset Background: T': 6762,
'KS Dataset Background: Y': 7068,
'KS Dataset Background: ST': 22743,
'KS Dataset Background: STY': 29811,
'Human Phosphoproteome Background: S': 86995,
'Human Phosphoproteome Background: T': 25091,
'Human Phosphoproteome Background: Y': 7869,
'Human Phosphoproteome Background: ST': 112086,
'Human Phosphoproteome Background: STY': 119955}plot_logos(bg_pssms_human,cnt,prefix='')
save_pdf('fig/background_human.pdf')