from katlas.core import *
from katlas.plot import *
import pandas as pd,numpy as np
from matplotlib import pyplot as pltScoring evaluation
Functions
def split_data(df):
df_st = df[df.kinase_group !='TK'].copy().reset_index(drop=True)
df_tyr = df[df.kinase_group =='TK'].copy().reset_index(drop=True)
df_st = df_st[df_st.site_seq.str[20]!='y'].copy().reset_index(drop=True)
df_tyr = df_tyr[df_tyr.site_seq.str[20]=='y'].copy().reset_index(drop=True)
return df_st, df_tyrdef split_ref(ref):
ref_st = ref[ref.index.isin(ST)].copy()
ref_tk = ref[ref.index.isin(TK)].copy()
return ref_st,ref_tkdef get_kinase_rank(row_index,df,result):
kinase = df.loc[row_index, 'kinase_protein']
scores = result.loc[row_index]
ranked = scores.sort_values(ascending=False)
rank = ranked.index.get_loc(kinase) + 1 # +1 to make rank start from 1
return rankdef score_data(df,ref,seq_col='site_seq',func=sumup,pct_ref=None):
df_st, df_tyr = split_data(df)
ref_st,ref_tk = split_ref(ref)
df_st_result = predict_kinase_df(df_st,seq_col=seq_col,ref=ref_st,func=func)
df_tyr_result = predict_kinase_df(df_tyr,seq_col=seq_col,ref=ref_tk,func=func)
if pct_ref is not None:
df_st_result = get_pct_df(df_st_result,pct_ref)
df_tyr_result = get_pct_df(df_tyr_result,pct_ref)
df_st['rank'] = df_st_result.index.to_series().apply(lambda idx: get_kinase_rank(idx,df_st,df_st_result))
df_tyr['rank'] = df_tyr_result.index.to_series().apply(lambda idx: get_kinase_rank(idx,df_tyr,df_tyr_result))
return df_st,df_tyr,df_st_result,df_tyr_resultdef get_ref(df, pssm, seq_col='site_seq', func=sumup, n_chunk=5):
chunk_size = (len(df) + n_chunk - 1) // n_chunk # ceiling division
pct_refs = []
for i in range(n_chunk):
start = i * chunk_size
end = min((i + 1) * chunk_size, len(df))
if start >= end:
break
print(f"Processing chunk {i+1}/{n_chunk} → rows {start}:{end}")
sub_df = df.iloc[start:end]
pct_ref = predict_kinase_df(
sub_df,
seq_col=seq_col,
ref=pssm,
func=func
)
pct_refs.append(pct_ref)
return pd.concat(pct_refs, ignore_index=True)def top_k_accuracy(df, result, k):
def is_in_top_k(row_index):
kinase = df.loc[row_index, 'kinase_protein']
scores = result.loc[row_index]
top_k = scores.nlargest(k).index
return kinase in top_k
return result.index.to_series().apply(is_in_top_k).mean()def top_k_accuracy_group(group_indices,df,result, k=10):
def is_correct(row_index):
kinase = df.loc[row_index, 'kinase_protein']
scores = result.loc[row_index]
top_k = scores.nlargest(k).index
return kinase in top_k
return pd.Series(group_indices).apply(is_correct).mean()def get_topk(df,result,k=10):
grouped = df.groupby('kinase_group').groups
topk_scores = {
subfam: top_k_accuracy_group(indices,df,result,k=k)
for subfam, indices in grouped.items()}
topk_df = pd.DataFrame.from_dict(topk_scores, orient='index', columns=['top10_accuracy']).sort_values('top10_accuracy', ascending=False)
return topk_dfdef get_group_topk(df_st,df_tyr,df_st_result,df_tyr_result):
topk_st = get_topk(df_st,df_st_result)
topk_tyr = get_topk(df_tyr,df_tyr_result)
topk = pd.concat([topk_st,topk_tyr]).sort_values('top10_accuracy',ascending=False)
return topkdef get_results(df_st,df_tyr,df_st_result,df_tyr_result):
topk = get_group_topk(df_st,df_tyr,df_st_result,df_tyr_result)
aucdf_st = get_AUCDF(df_st,'rank')
aucdf_tyr = get_AUCDF(df_tyr,'rank')
return topk, aucdf_st,aucdf_tyrset_sns()Overall
pspa
pspa = pd.read_parquet('raw/overlap_pspa.parquet')
pspa.index = pspa.index.str.split('_').str[1]
pspa.shape(312, 236)Data to be scored
psp = Data.get_ks_dataset()df = psp[psp.source.str.contains('PSP')].copy()df['site_seq_upper'] = df.site_seq.str.upper()# only filter those with tested kinase
df = df[df.kinase_protein.isin(pspa.index)].copy().reset_index(drop=True)df.shape(12639, 22)group_map = df[['kinase_protein','kinase_group']].drop_duplicates().set_index('kinase_protein')['kinase_group']
TK = group_map[group_map=='TK'].index
ST = group_map[group_map!='TK'].indexdef prepare_ref(path):
ref = pd.read_parquet(path)
ref.index = ref.index.str.split('_').str[1]
ref = ref[ref.index.isin(pspa.index)]
print(ref.shape)
return refcddm:
cddm = prepare_ref('out/CDDM_pssms.parquet')
cddm_upper = prepare_ref('out/CDDM_pssms_upper.parquet')
cddm_lo = prepare_ref('out/CDDM_pssms_LO.parquet')
cddm_lo_upper = prepare_ref('out/CDDM_pssms_LO_upper.parquet')(312, 943)
(312, 943)
(312, 943)
(312, 943)# ref_dict = {
# 'PSPA: PSSM + multiply': (pspa,'site_seq',multiply,False),
# 'CDDM: PSSM + multiply': (cddm, 'site_seq', multiply_23,False),
# 'CDDM: PSSM upper + multiply': (cddm_upper, 'site_seq_upper', multiply_20,False),
# 'CDDM: LO + sum': (cddm_lo, 'site_seq',sumup,False),
# 'CDDM: LO upper + sum': (cddm_lo_upper, 'site_seq_upper',sumup,False),
# }
# human =Data.get_human_site()
# human['site_seq_upper'] = human.site_seq.str.upper()
# human = human.drop_duplicates('site_seq').reset_index()
# for name,(ref,seq_col,func,_) in ref_dict.items():
# print(name)
# pct_ref = get_ref(human,pssm=ref,seq_col=seq_col,func=func)
# pct_ref.to_parquet(f'raw/{name}.parquet')
# print(pct_ref.head())
# # breakref_dict = {
'PSPA: PSSM + multiply': (pspa,'site_seq',multiply,False),
'PSPA: PSSM + multiply + pct': (pspa,'site_seq',multiply, True),
'CDDM: PSSM + multiply': (cddm, 'site_seq', multiply_23,False),
'CDDM: PSSM + multiply + pct': (cddm, 'site_seq', multiply_23, True),
'CDDM: PSSM upper + multiply': (cddm_upper, 'site_seq_upper', multiply_20,False),
'CDDM: PSSM upper + multiply + pct': (cddm_upper, 'site_seq_upper', multiply_20,True),
'CDDM: LO + sum': (cddm_lo, 'site_seq',sumup,False),
'CDDM: LO + sum + pct': (cddm_lo, 'site_seq',sumup,True),
'CDDM: LO upper + sum': (cddm_lo_upper, 'site_seq_upper',sumup,False),
'CDDM: LO upper + sum + pct': (cddm_lo_upper, 'site_seq_upper',sumup,True),
}topk_dfs = []
aucdf = pd.DataFrame()
for name,(ref,seq_col,func,use_pct) in ref_dict.items():
print('---------------------------------------------')
print(name)
pct_ref = pd.read_parquet(f'raw/{name[:-6]}.parquet') if use_pct else None # -6 to remove + pct from parquet name
df_st,df_tyr,df_st_result,df_tyr_result = score_data(df,ref,seq_col=seq_col,func=func,pct_ref=pct_ref)
topk,aucdf_st,aucdf_tyr = get_results(df_st,df_tyr,df_st_result,df_tyr_result)
topk_dfs.append(topk)
aucdf.loc[name,'ST'] = aucdf_st
aucdf.loc[name,'Tyr'] = aucdf_tyr---------------------------------------------
PSPA: PSSM + multiply
input dataframe has a length 10603
Preprocessing
Finish preprocessing
Merging reference
Finish merging100%|███████████████████████████████████████████████████████████████████████████| 213/213 [00:02<00:00, 81.69it/s]input dataframe has a length 1912
Preprocessing
Finish preprocessing
Merging reference
Finish merging100%|████████████████████████████████████████████████████████████████████████████| 71/71 [00:00<00:00, 428.49it/s]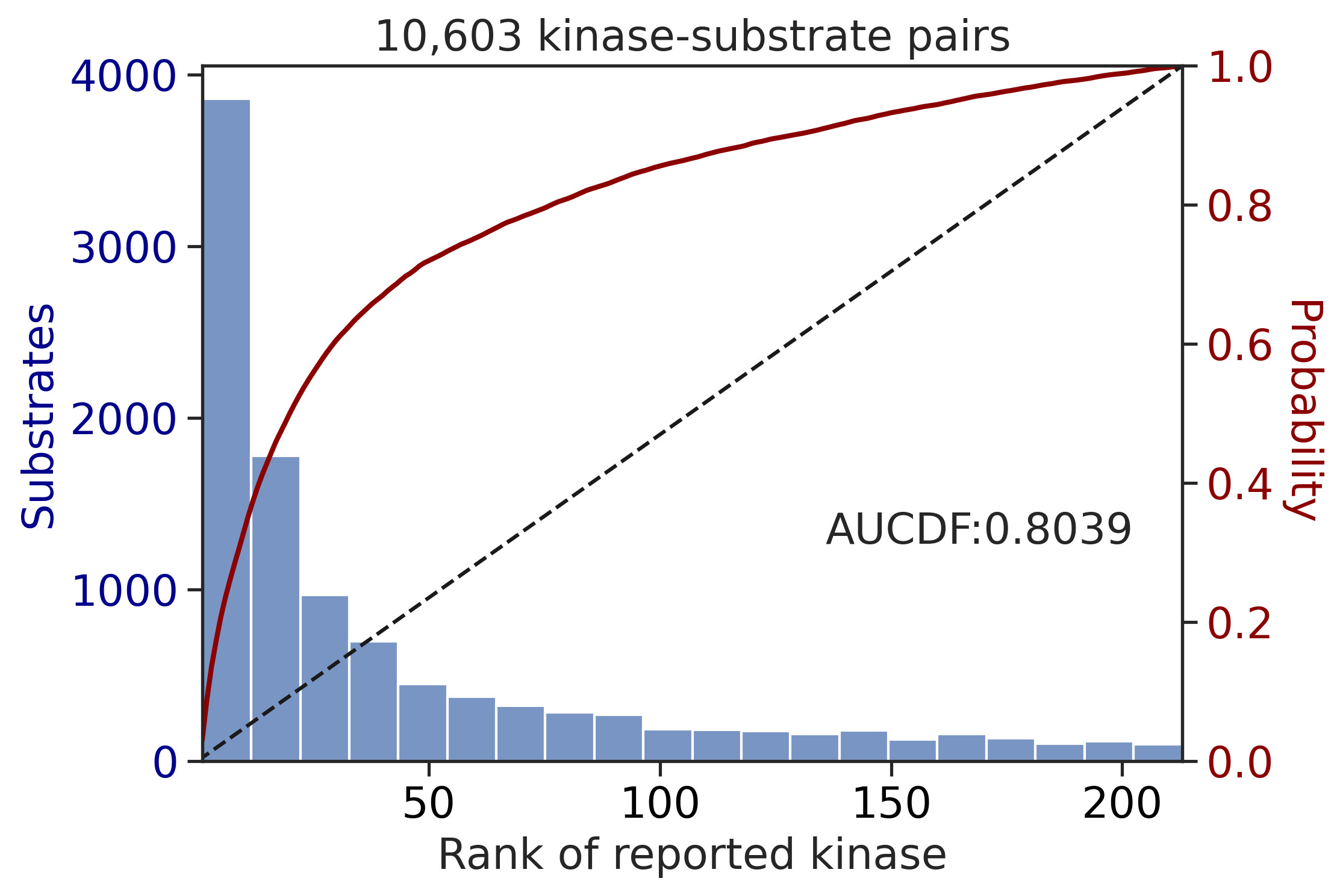
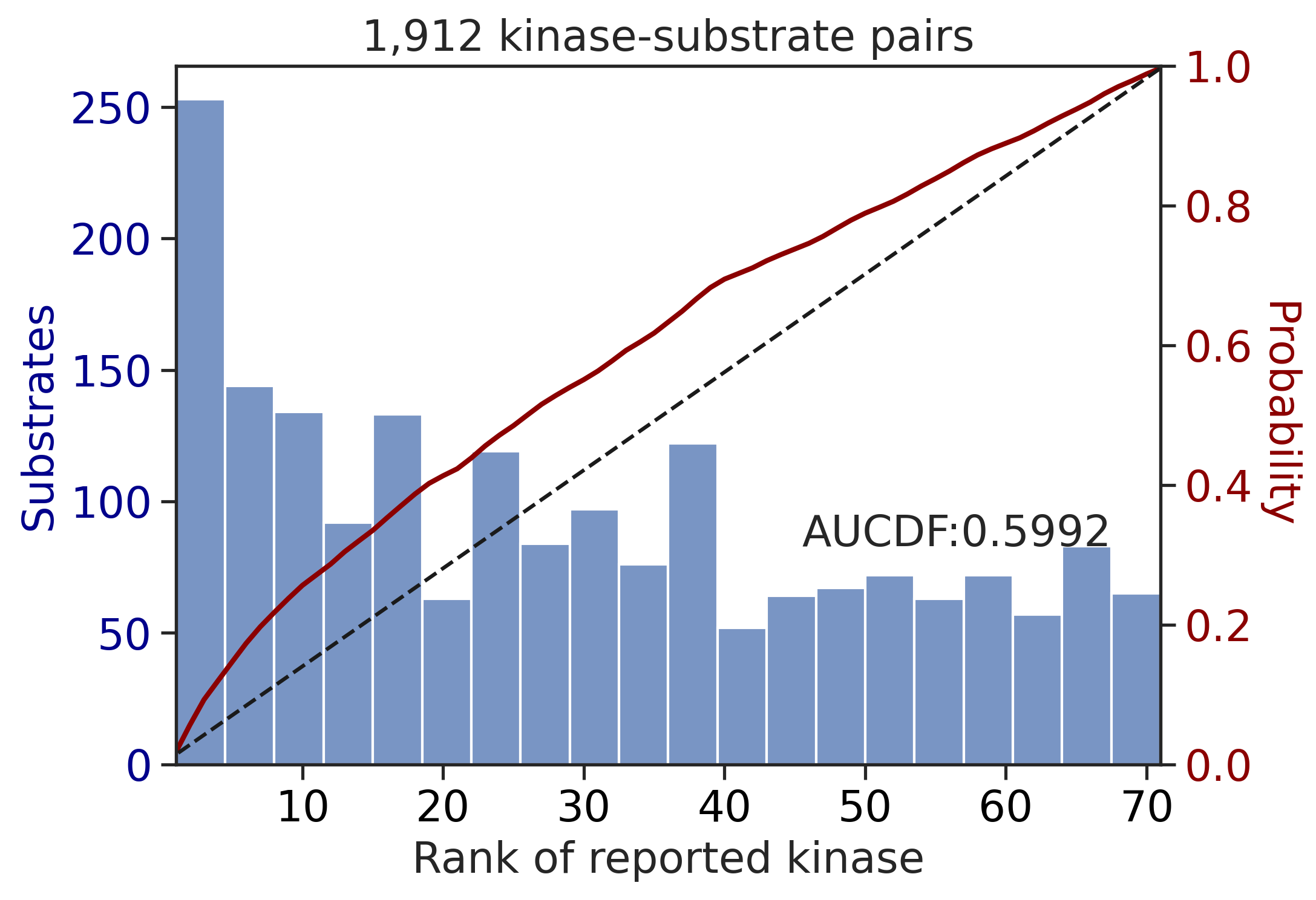
---------------------------------------------
PSPA: PSSM + multiply + pct
input dataframe has a length 10603
Preprocessing
Finish preprocessing
Merging reference
Finish merging100%|███████████████████████████████████████████████████████████████████████████| 213/213 [00:02<00:00, 85.26it/s]input dataframe has a length 1912
Preprocessing
Finish preprocessing
Merging reference
Finish merging100%|████████████████████████████████████████████████████████████████████████████| 71/71 [00:00<00:00, 423.93it/s]
100%|██████████████████████████████████████████████████████████████████████████| 213/213 [00:00<00:00, 389.86it/s]
100%|████████████████████████████████████████████████████████████████████████████| 71/71 [00:00<00:00, 764.05it/s]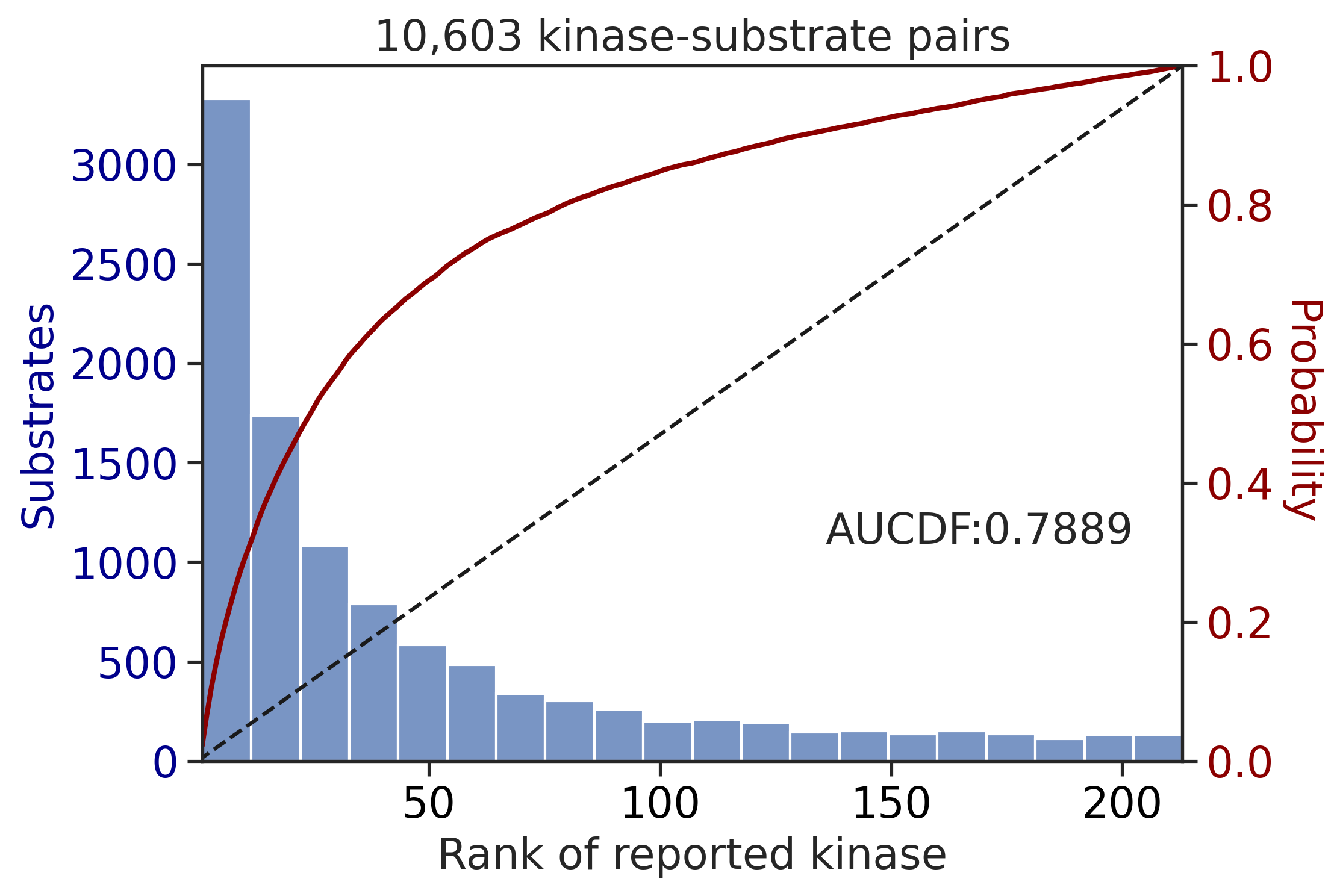
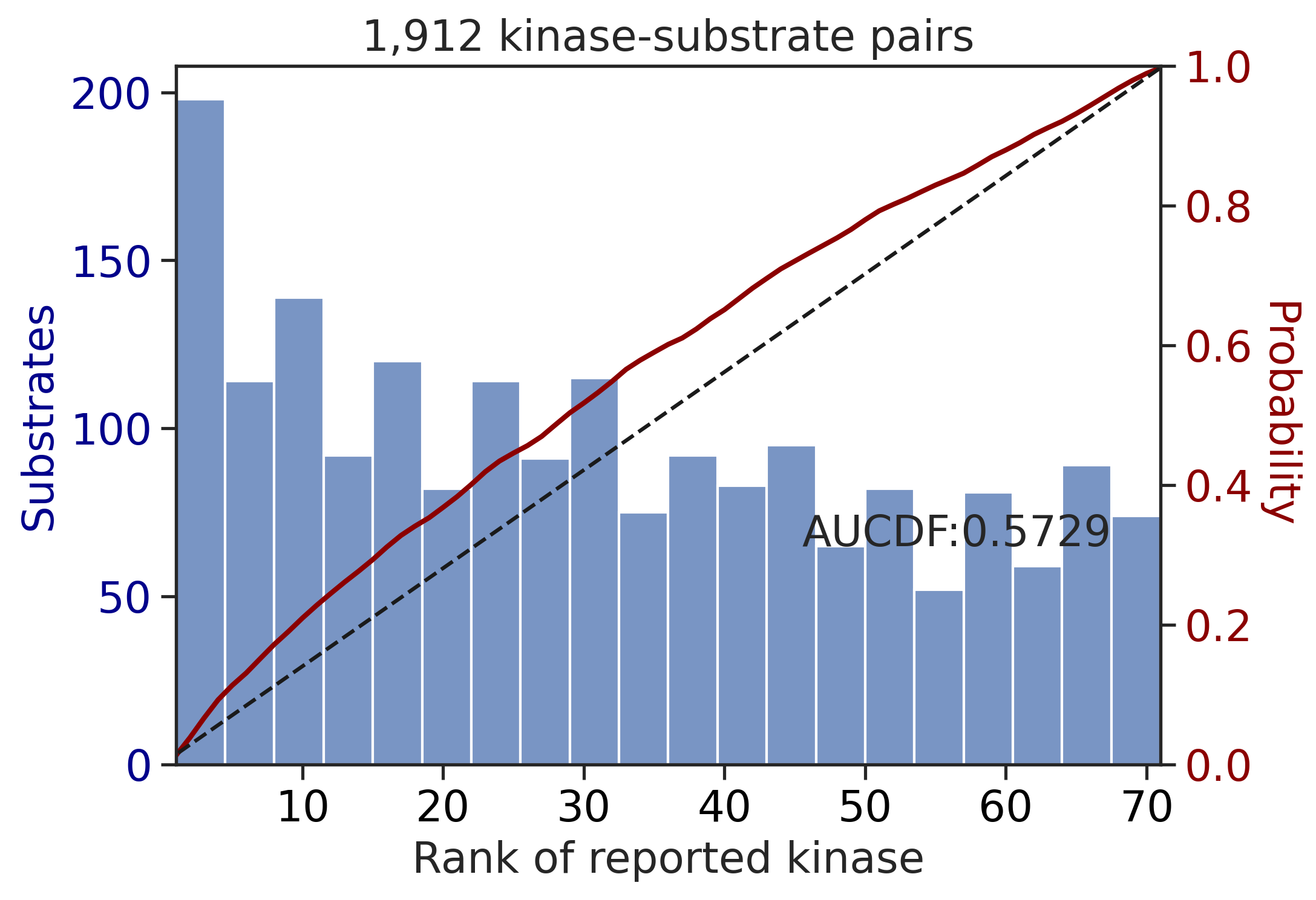
---------------------------------------------
CDDM: PSSM + multiply
input dataframe has a length 10603
Preprocessing
Finish preprocessing
Merging reference
Finish merging100%|███████████████████████████████████████████████████████████████████████████| 213/213 [00:04<00:00, 50.58it/s]input dataframe has a length 1912
Preprocessing
Finish preprocessing
Merging reference
Finish merging100%|████████████████████████████████████████████████████████████████████████████| 71/71 [00:00<00:00, 215.43it/s]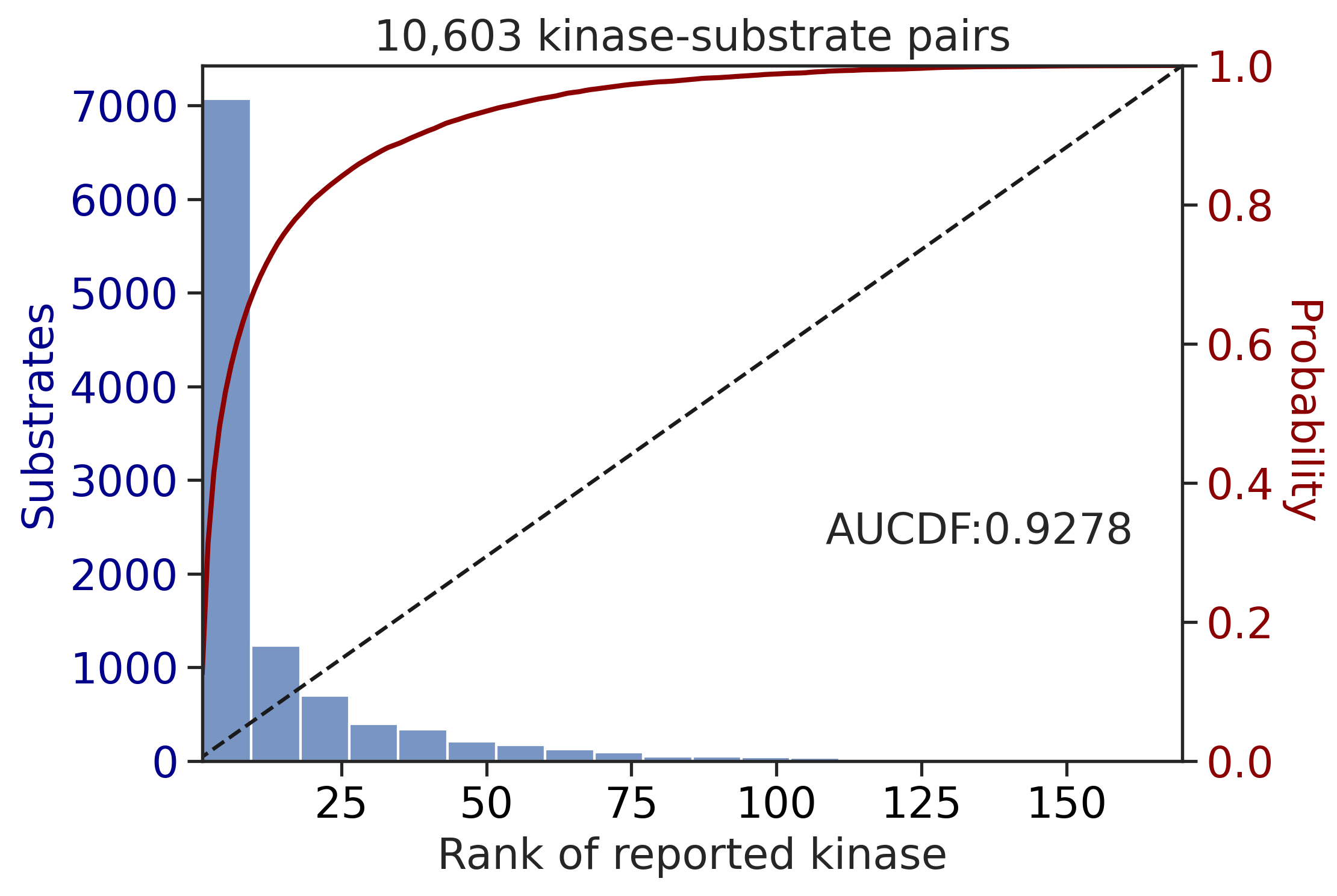
---------------------------------------------
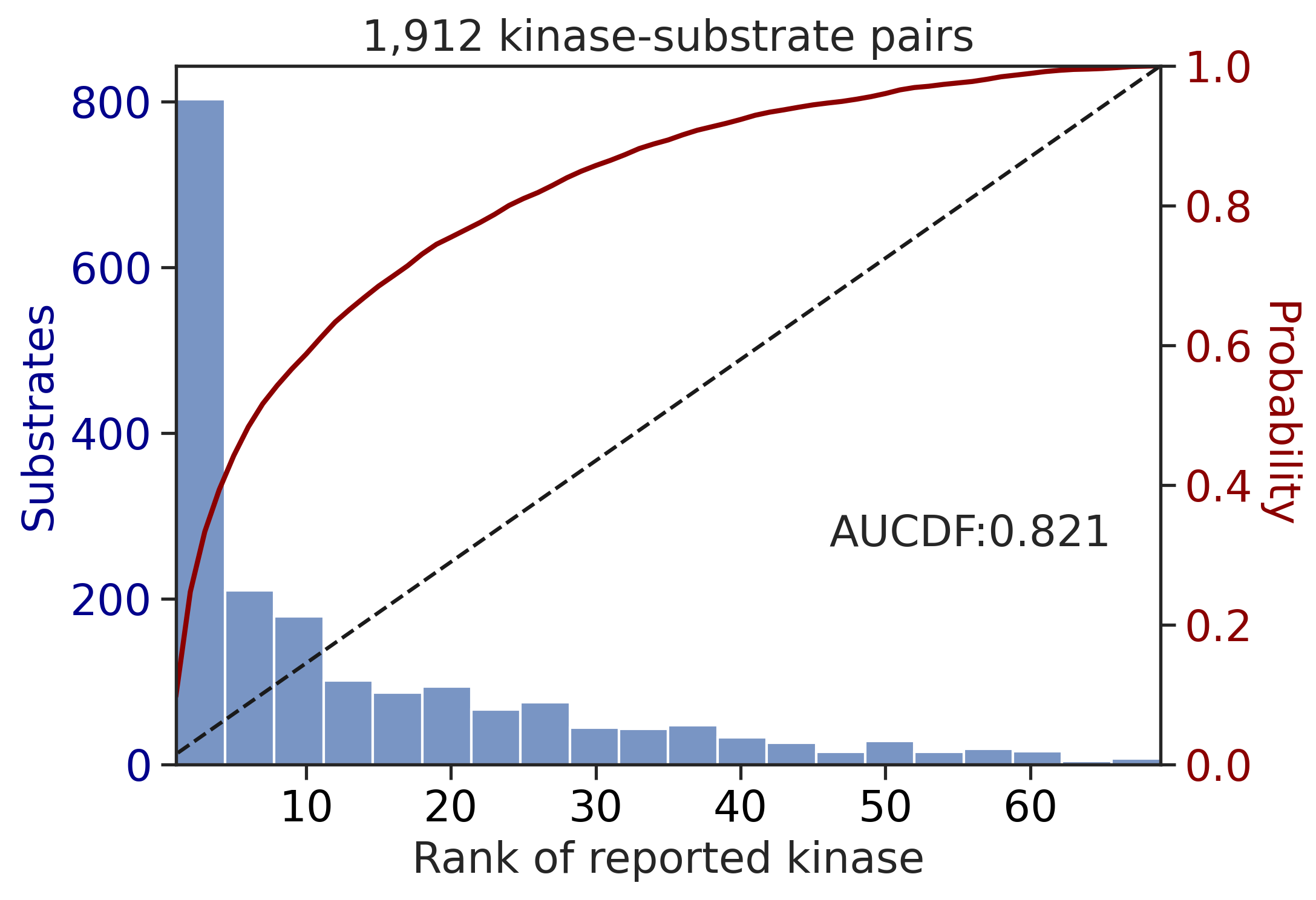
CDDM: PSSM + multiply + pct
input dataframe has a length 10603
Preprocessing
Finish preprocessing
Merging reference
Finish merging100%|███████████████████████████████████████████████████████████████████████████| 213/213 [00:04<00:00, 50.47it/s]input dataframe has a length 1912
Preprocessing
Finish preprocessing
Merging reference
Finish merging100%|████████████████████████████████████████████████████████████████████████████| 71/71 [00:00<00:00, 177.45it/s]
100%|██████████████████████████████████████████████████████████████████████████| 213/213 [00:00<00:00, 373.54it/s]
100%|████████████████████████████████████████████████████████████████████████████| 71/71 [00:00<00:00, 509.36it/s]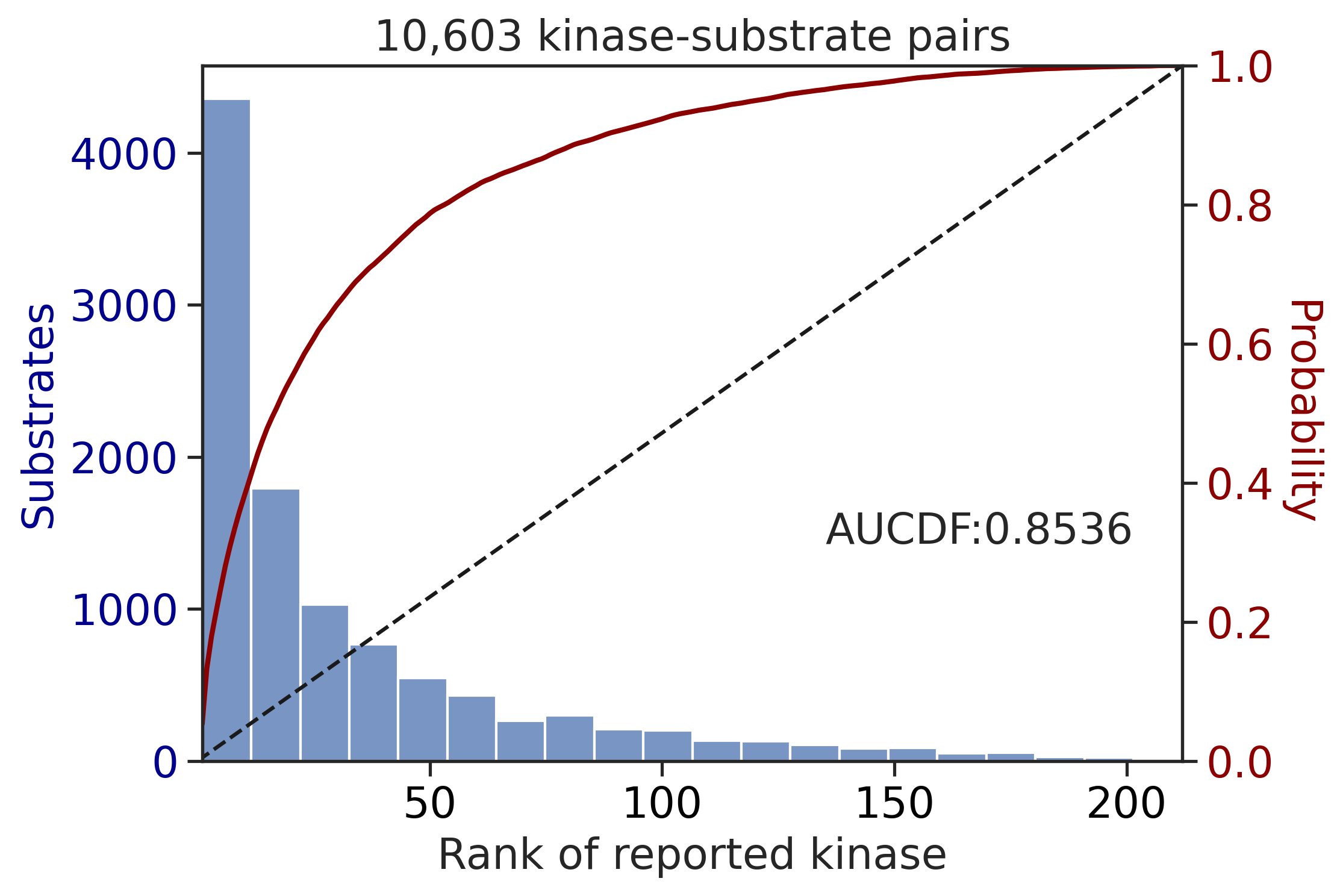
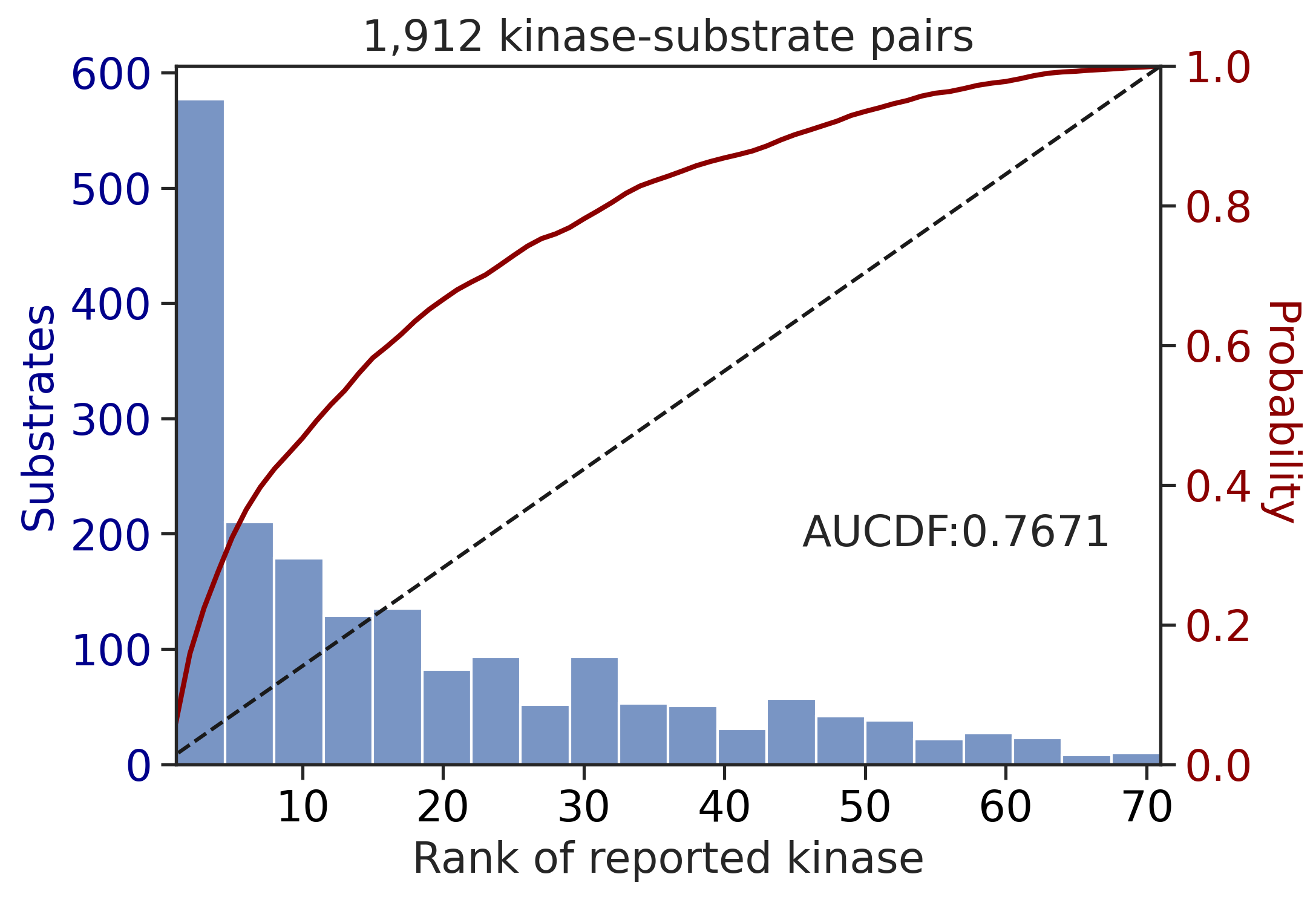
---------------------------------------------
CDDM: PSSM upper + multiply
input dataframe has a length 10603
Preprocessing
Finish preprocessing
Merging reference
Finish merging100%|███████████████████████████████████████████████████████████████████████████| 213/213 [00:04<00:00, 51.73it/s]input dataframe has a length 1912
Preprocessing
Finish preprocessing
Merging reference
Finish merging100%|████████████████████████████████████████████████████████████████████████████| 71/71 [00:00<00:00, 228.69it/s]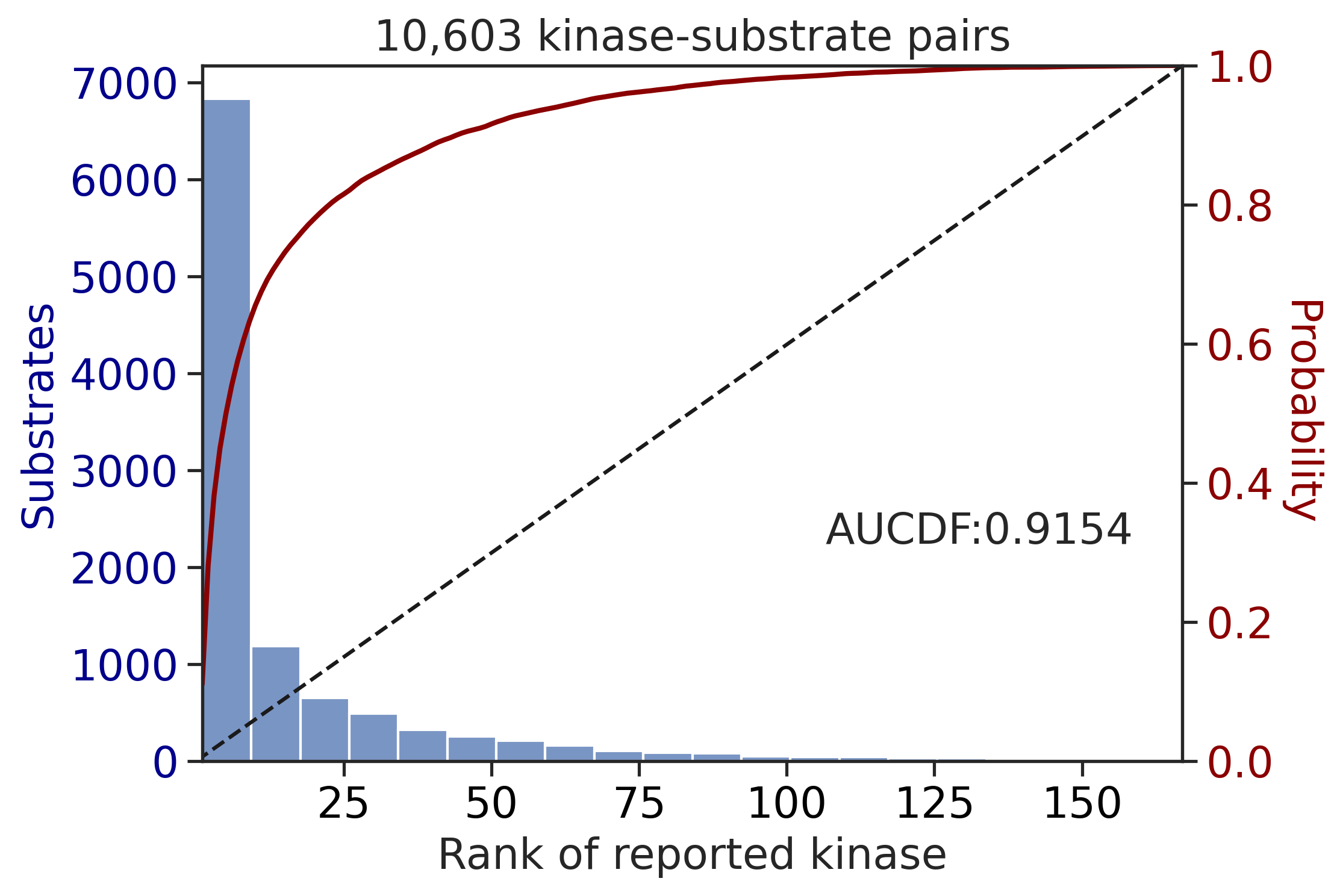
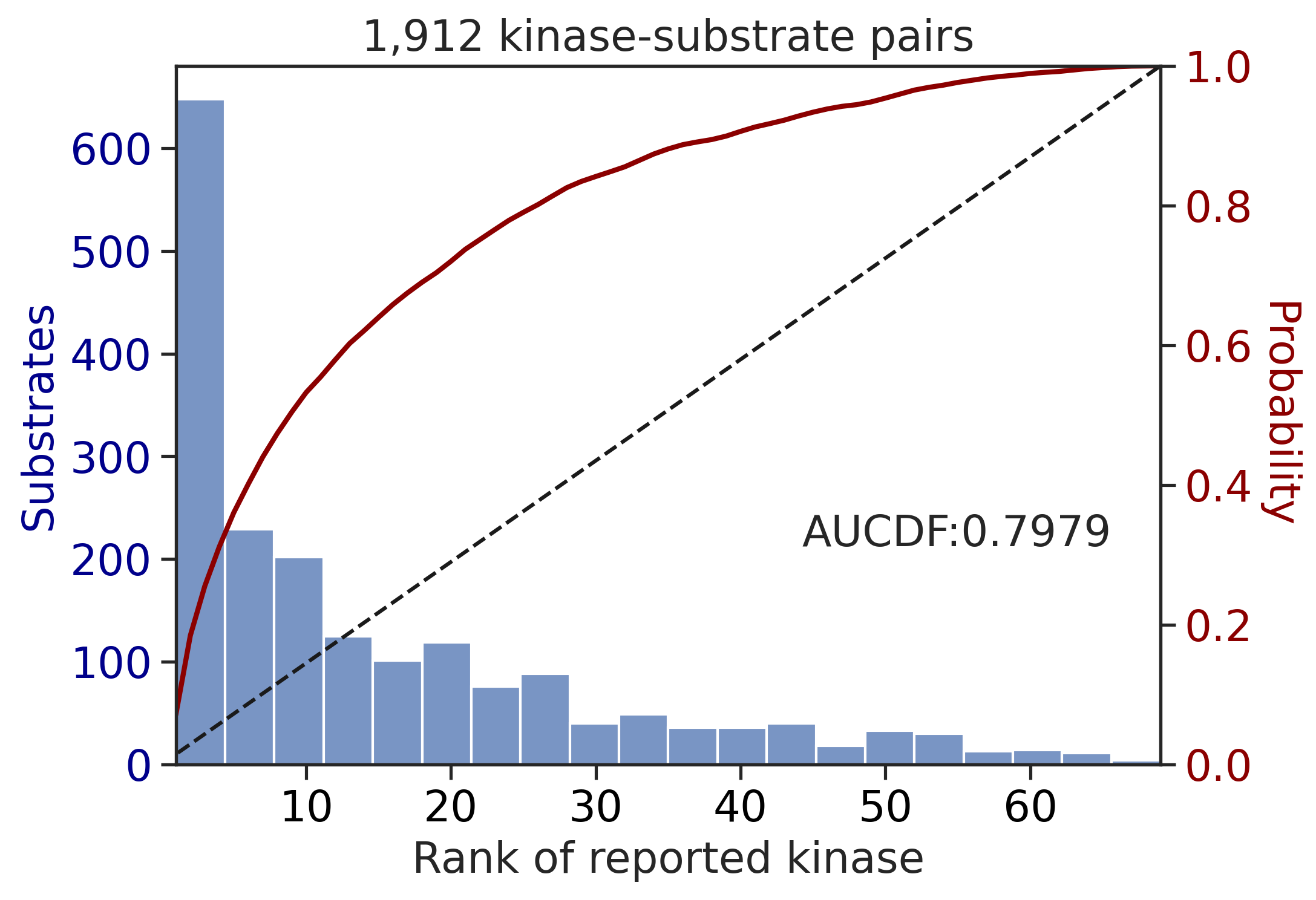
---------------------------------------------
CDDM: PSSM upper + multiply + pct
input dataframe has a length 10603
Preprocessing
Finish preprocessing
Merging reference
Finish merging100%|███████████████████████████████████████████████████████████████████████████| 213/213 [00:04<00:00, 52.55it/s]input dataframe has a length 1912
Preprocessing
Finish preprocessing
Merging reference
Finish merging100%|████████████████████████████████████████████████████████████████████████████| 71/71 [00:00<00:00, 225.45it/s]
100%|██████████████████████████████████████████████████████████████████████████| 213/213 [00:00<00:00, 388.72it/s]
100%|████████████████████████████████████████████████████████████████████████████| 71/71 [00:00<00:00, 701.91it/s]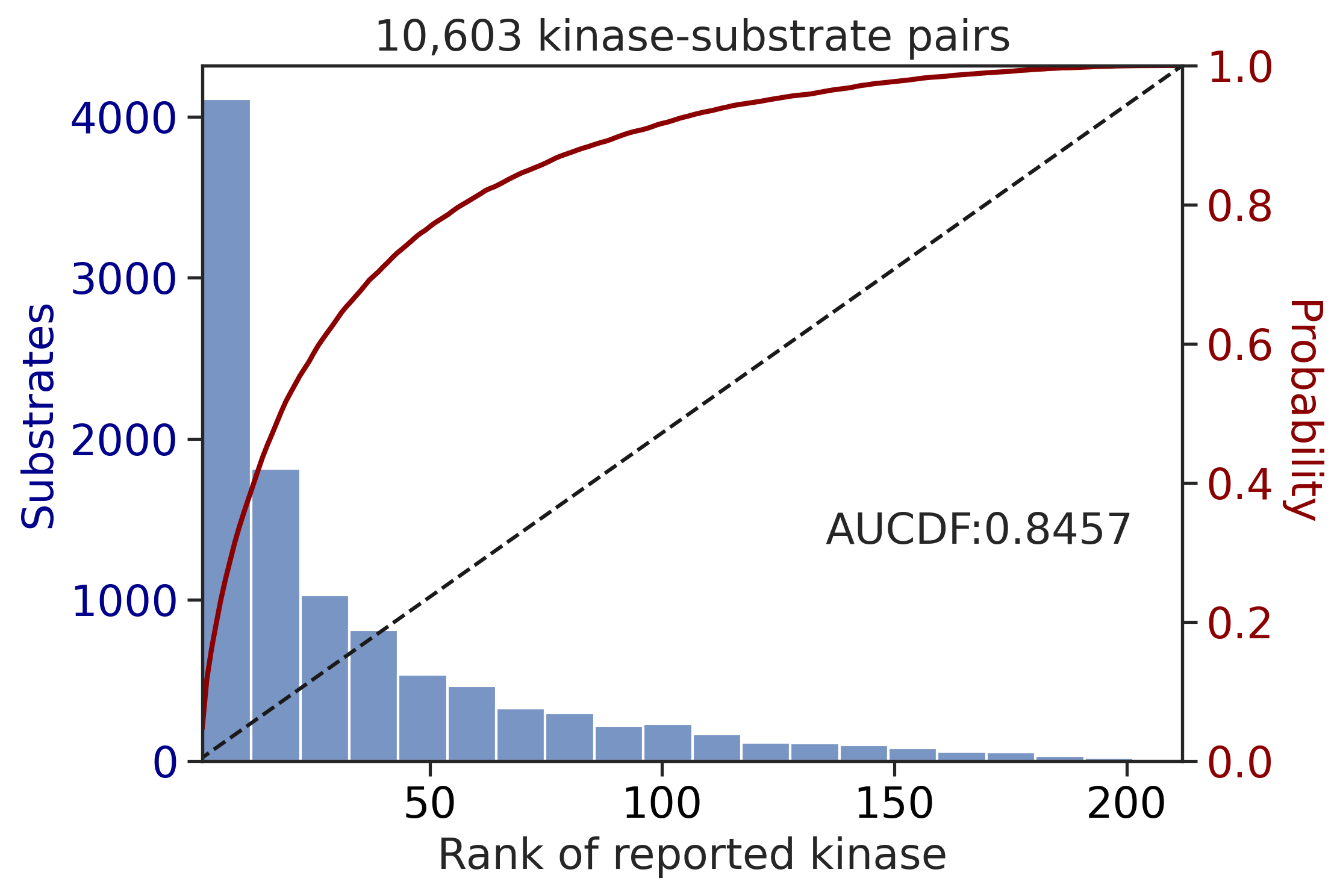
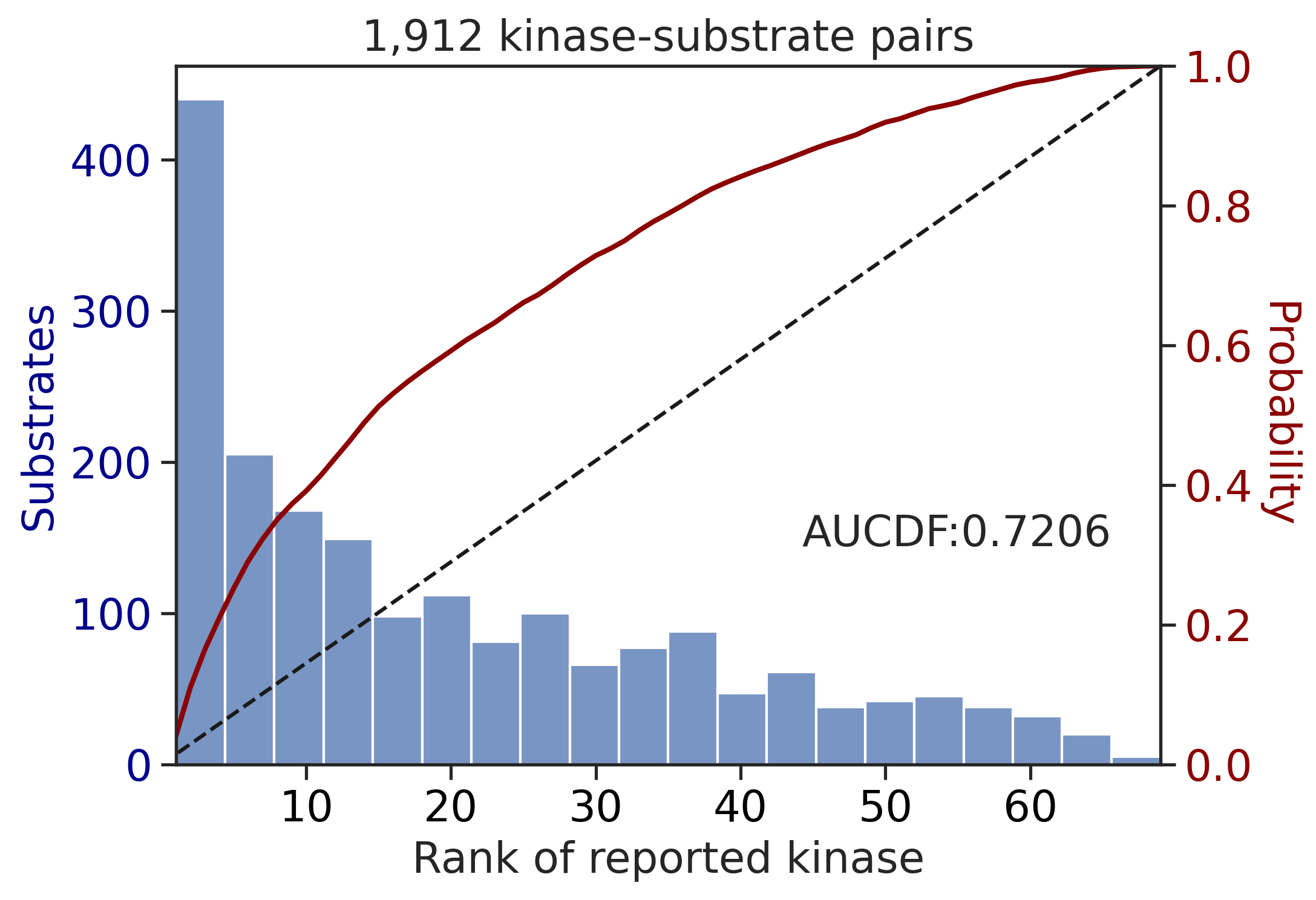
---------------------------------------------
CDDM: LO + sum
input dataframe has a length 10603
Preprocessing
Finish preprocessing
Merging reference
Finish merging
input dataframe has a length 1912
Preprocessing
Finish preprocessing
Merging reference
Finish merging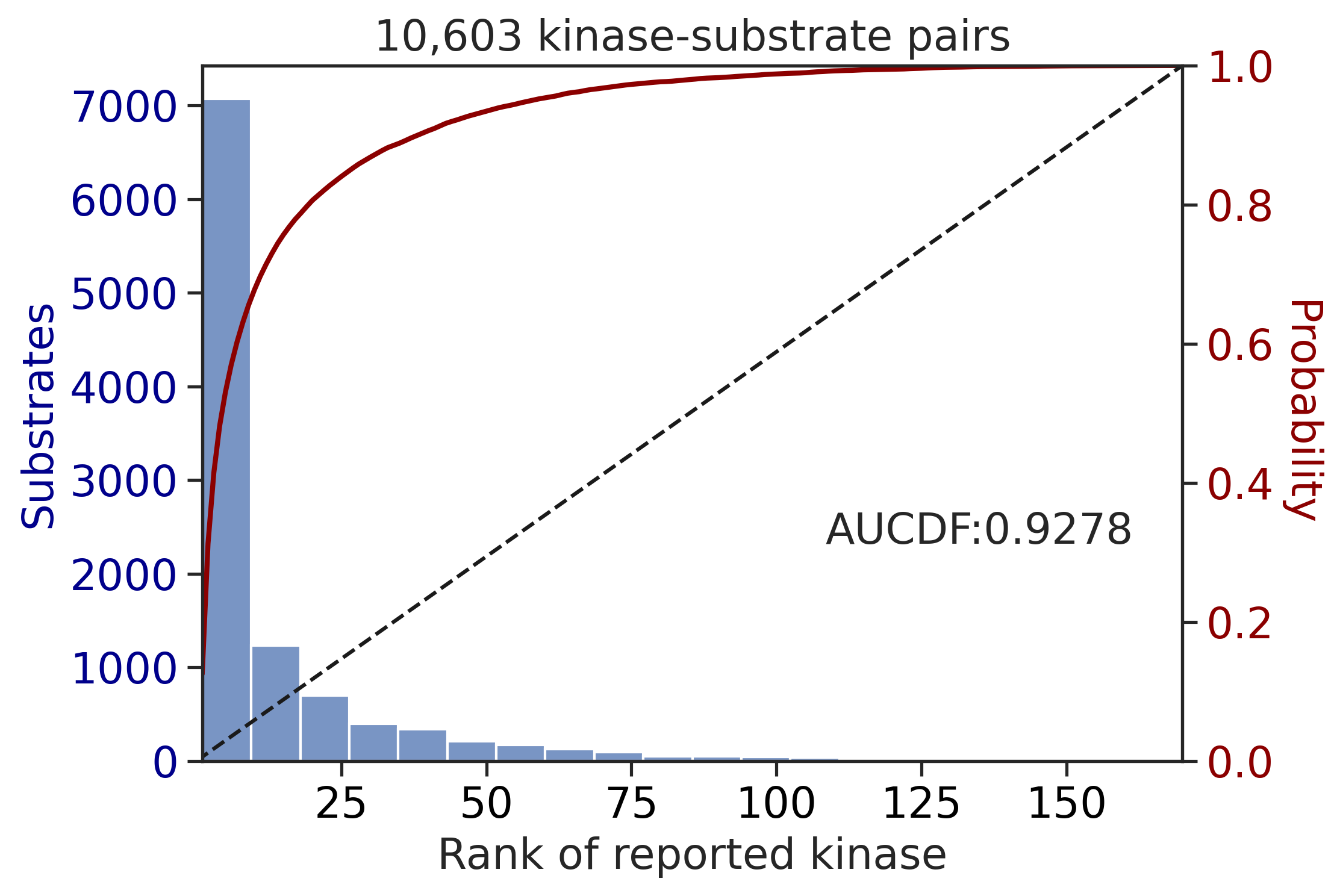
---------------------------------------------
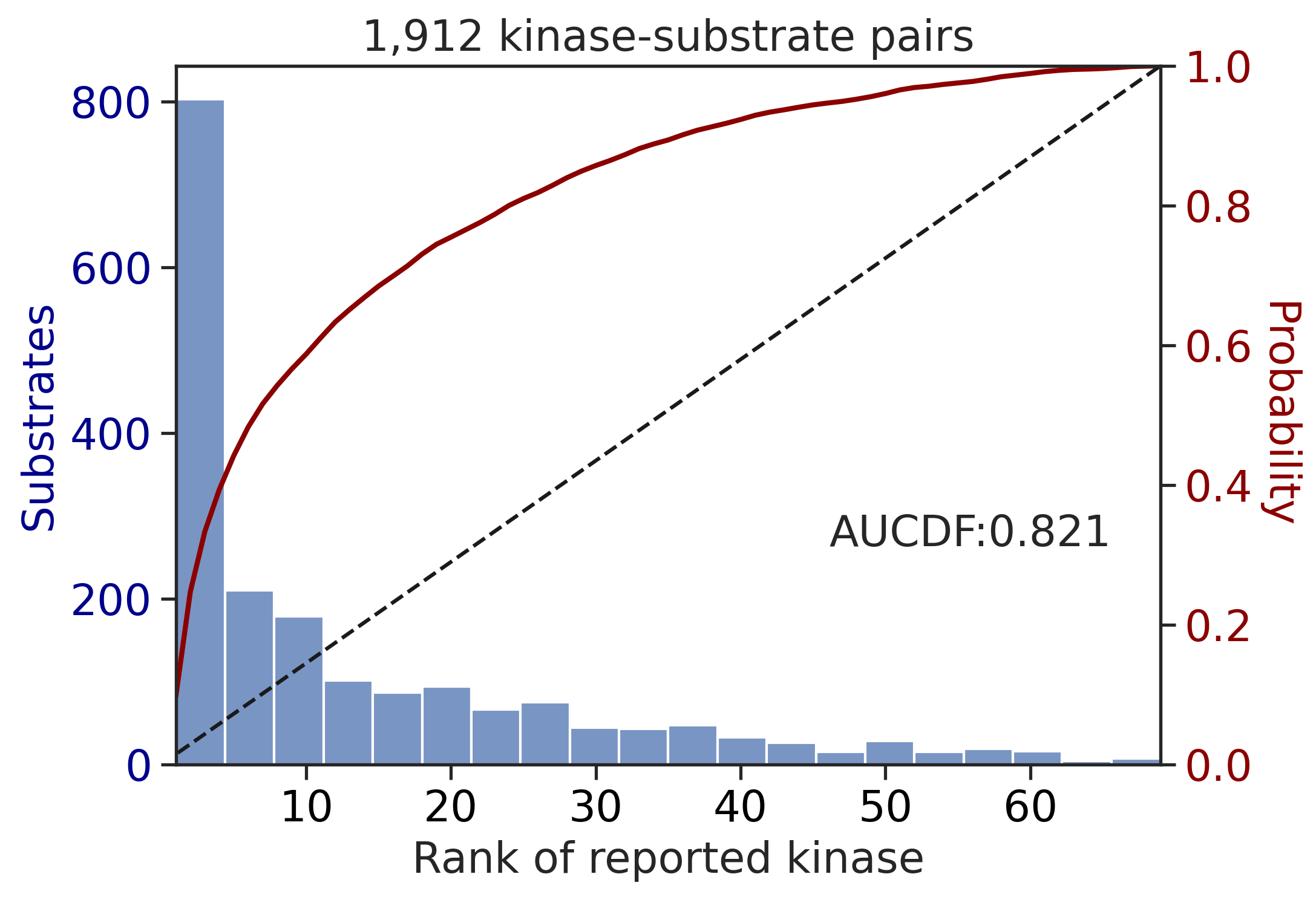
CDDM: LO + sum + pct
input dataframe has a length 10603
Preprocessing
Finish preprocessing
Merging reference
Finish merging
input dataframe has a length 1912
Preprocessing
Finish preprocessing
Merging reference
Finish merging100%|██████████████████████████████████████████████████████████████████████████| 213/213 [00:00<00:00, 382.47it/s]
100%|████████████████████████████████████████████████████████████████████████████| 71/71 [00:00<00:00, 749.55it/s]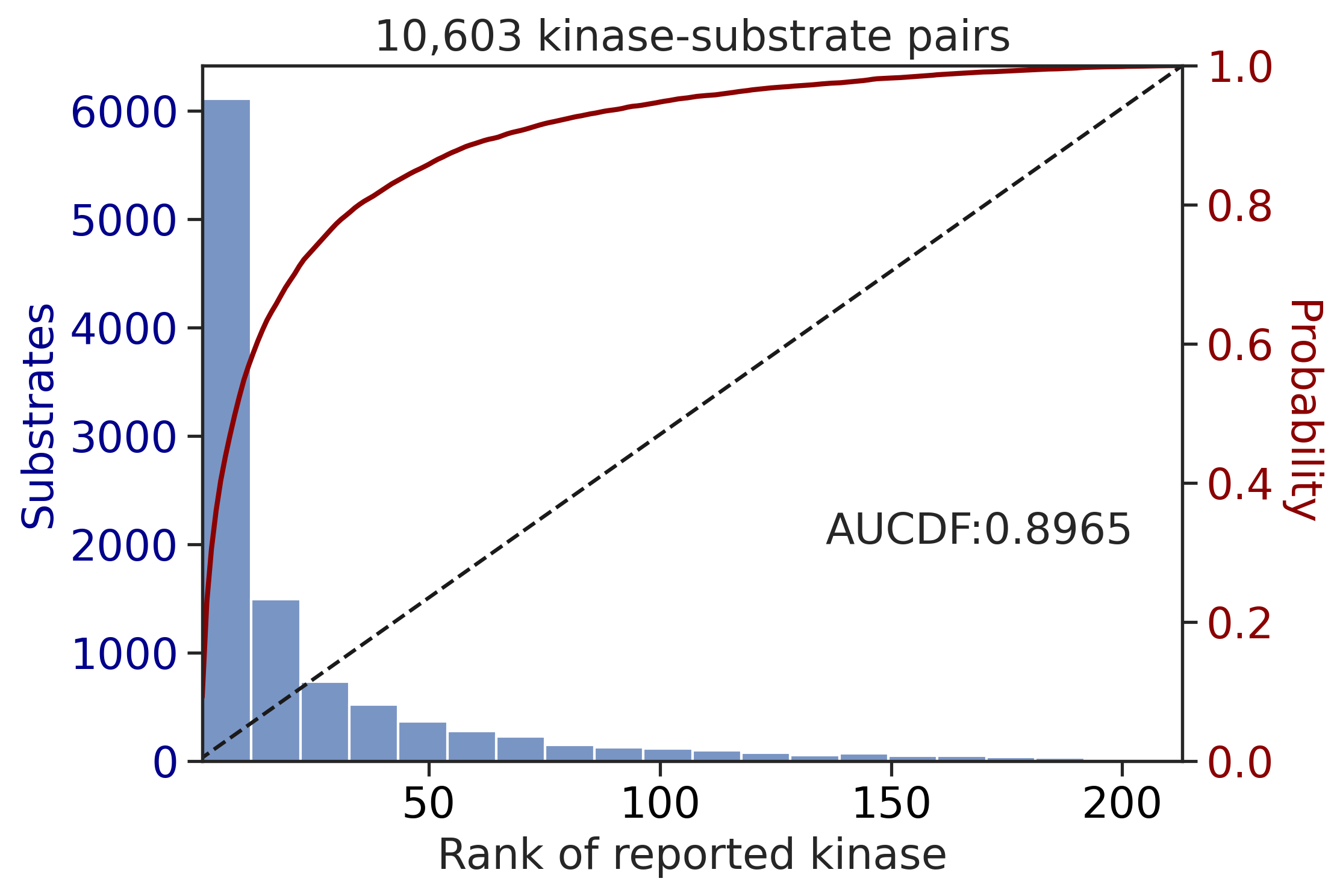
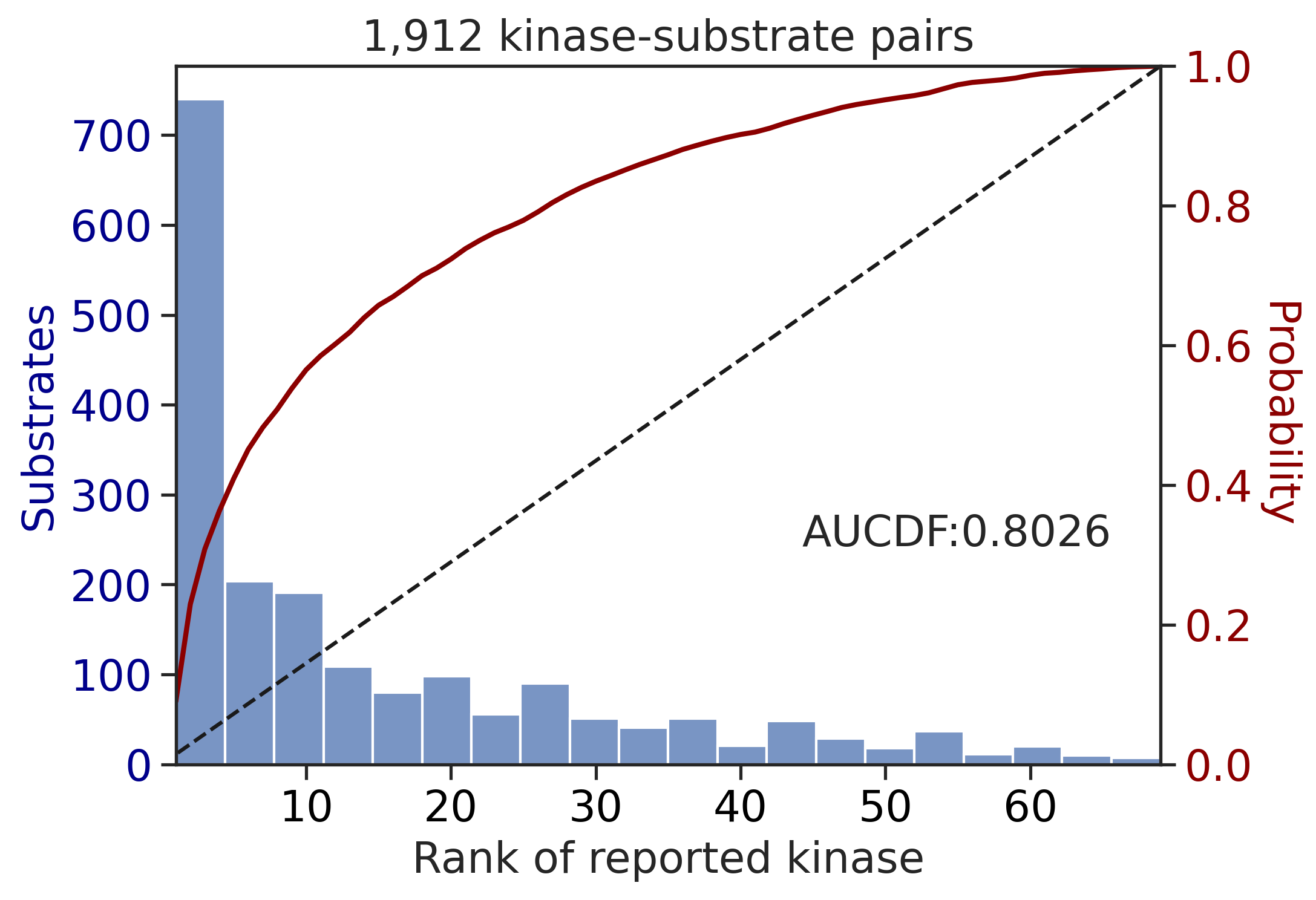
---------------------------------------------
CDDM: LO upper + sum
input dataframe has a length 10603
Preprocessing
Finish preprocessing
Merging reference
Finish merging
input dataframe has a length 1912
Preprocessing
Finish preprocessing
Merging reference
Finish merging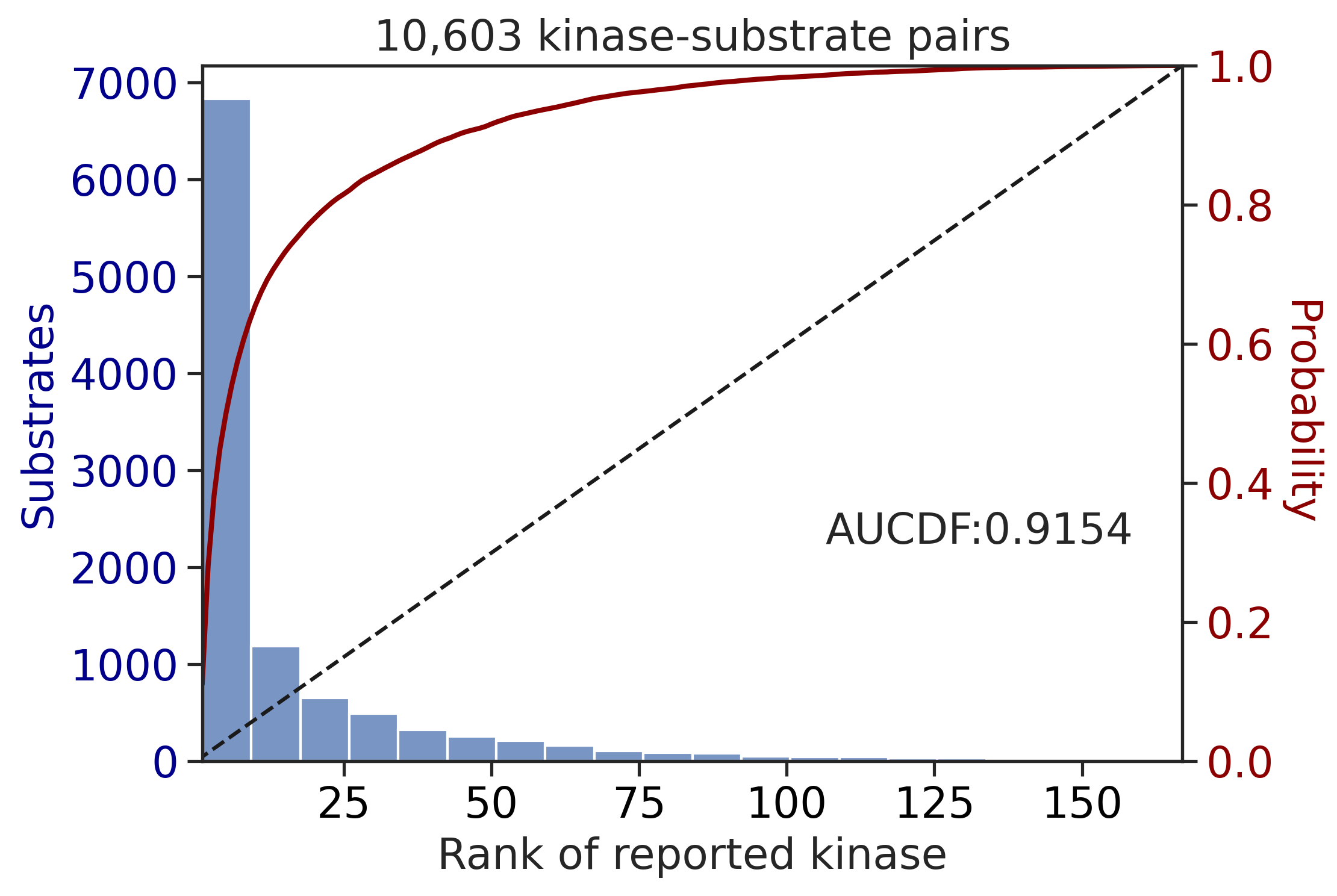
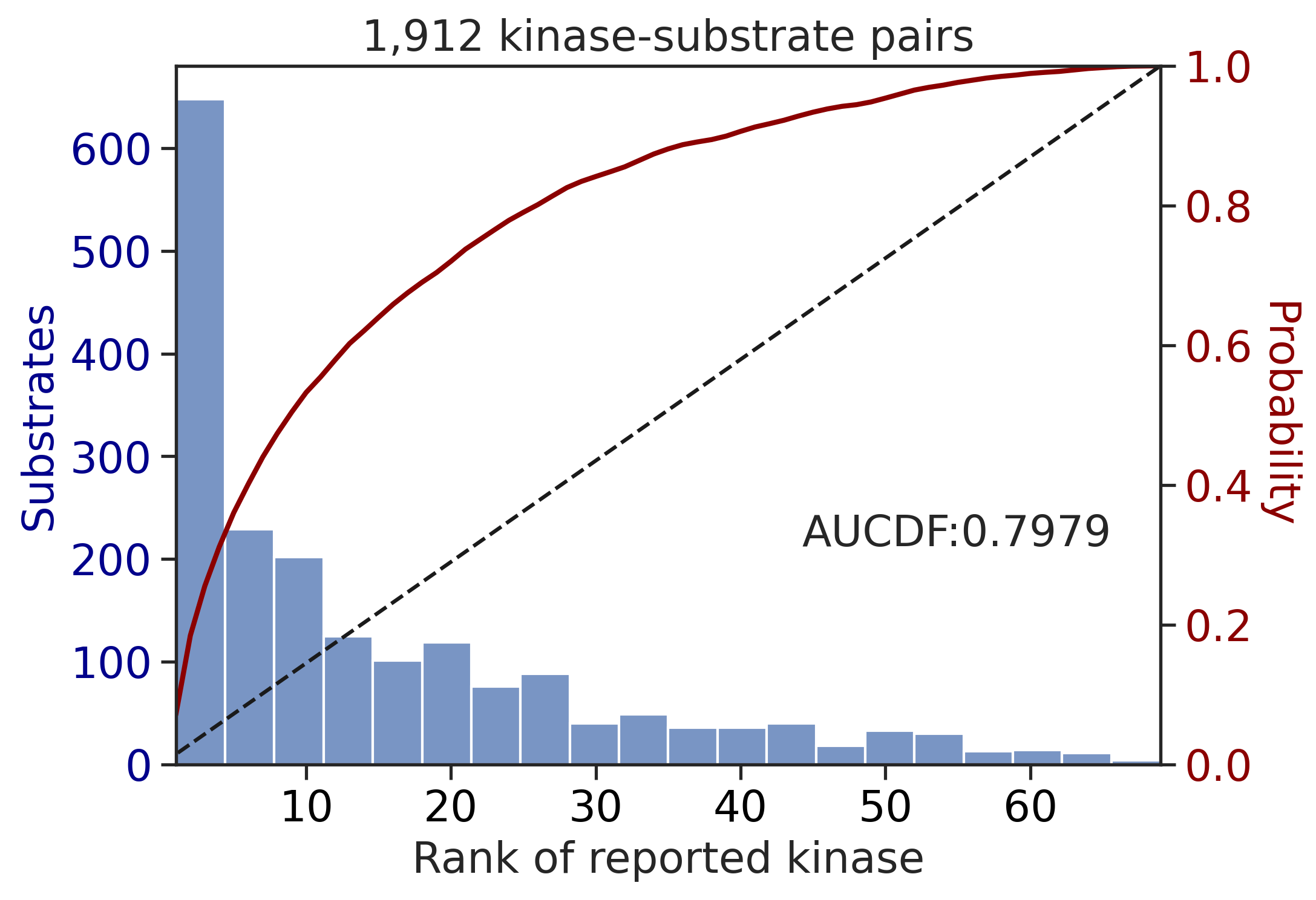
---------------------------------------------
CDDM: LO upper + sum + pct
input dataframe has a length 10603
Preprocessing
Finish preprocessing
Merging reference
Finish merging
input dataframe has a length 1912
Preprocessing
Finish preprocessing
Merging reference
Finish merging100%|██████████████████████████████████████████████████████████████████████████| 213/213 [00:00<00:00, 378.69it/s]
100%|████████████████████████████████████████████████████████████████████████████| 71/71 [00:00<00:00, 764.31it/s]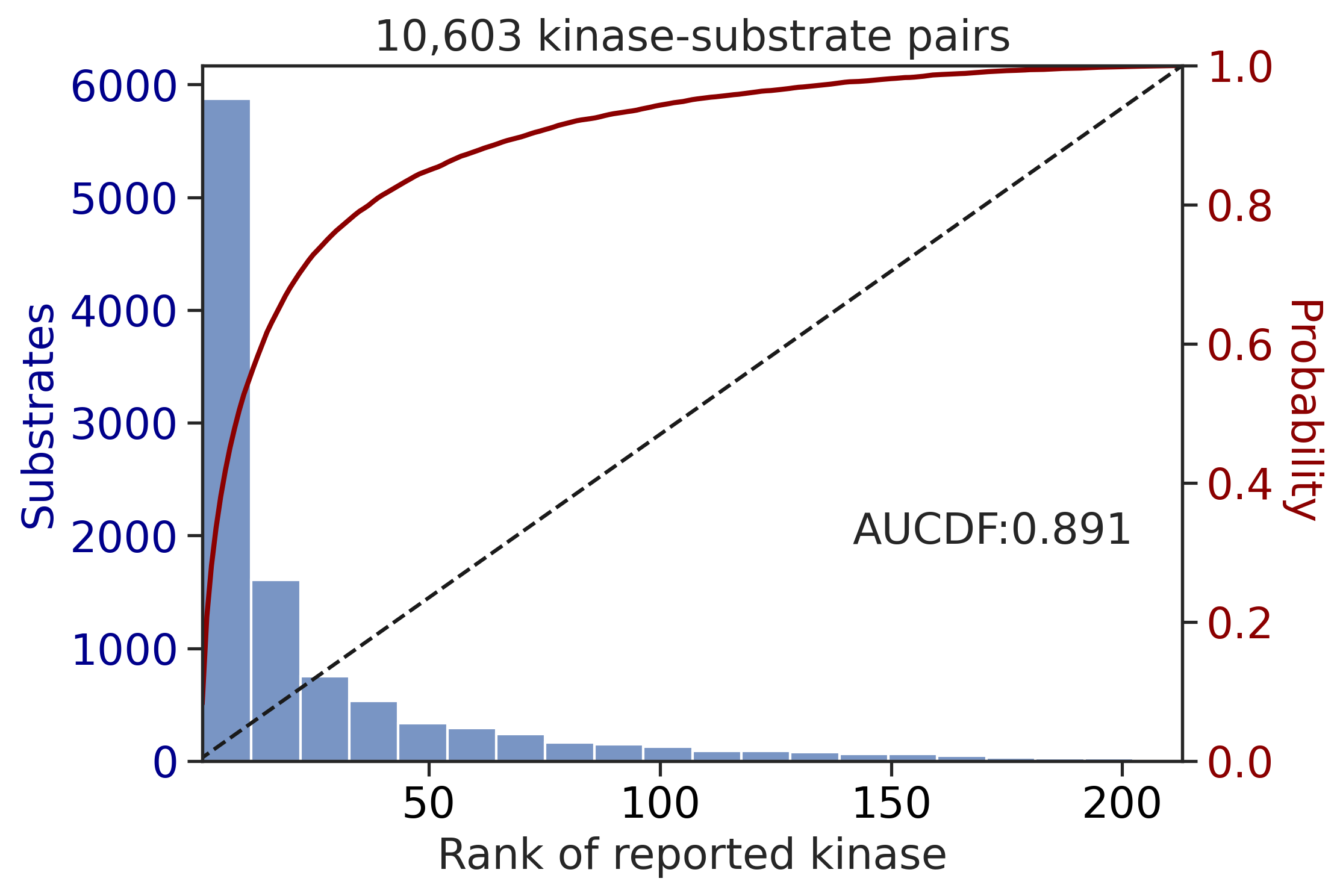
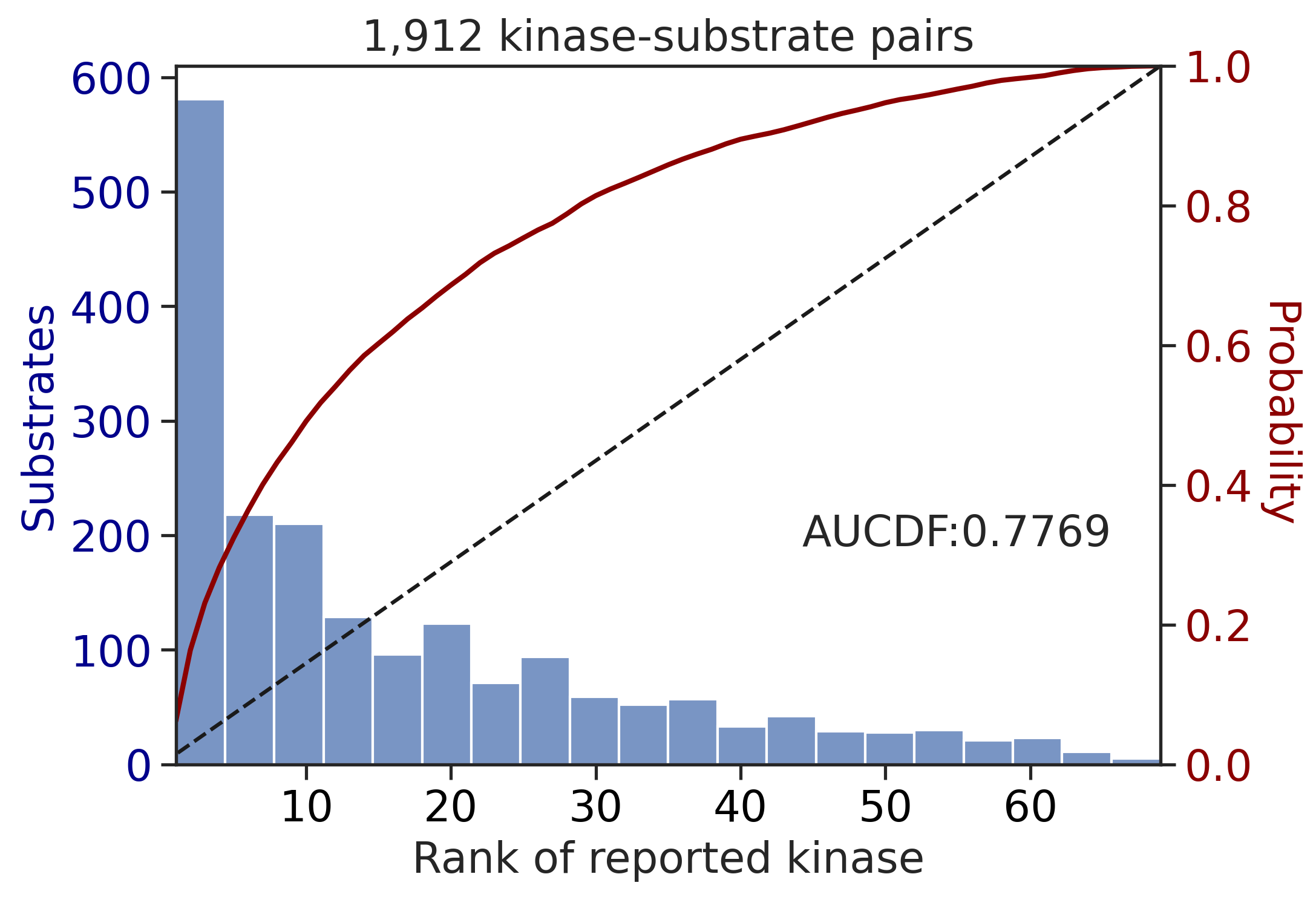
aucdf = aucdf.Taucdf = aucdf.reset_index(names='group')aucdf.set_index('group').T.sort_values('Tyr')| group | ST | Tyr |
|---|---|---|
| PSPA: PSSM + multiply + pct | 0.788949 | 0.572925 |
| PSPA: PSSM + multiply | 0.803919 | 0.599230 |
| CDDM: PSSM upper + multiply + pct | 0.845689 | 0.720623 |
| CDDM: PSSM + multiply + pct | 0.853593 | 0.767071 |
| CDDM: LO upper + sum + pct | 0.890996 | 0.776887 |
| CDDM: PSSM upper + multiply | 0.915436 | 0.797867 |
| CDDM: LO upper + sum | 0.915436 | 0.797867 |
| CDDM: LO + sum + pct | 0.896505 | 0.802644 |
| CDDM: LO + sum | 0.927760 | 0.821030 |
| CDDM: PSSM + multiply | 0.927760 | 0.821030 |
plot_group_bar(aucdf,value_cols=aucdf.columns[1:],group='group',rotation=0)
plt.ylabel('AUCDF')Text(0, 0.5, 'AUCDF')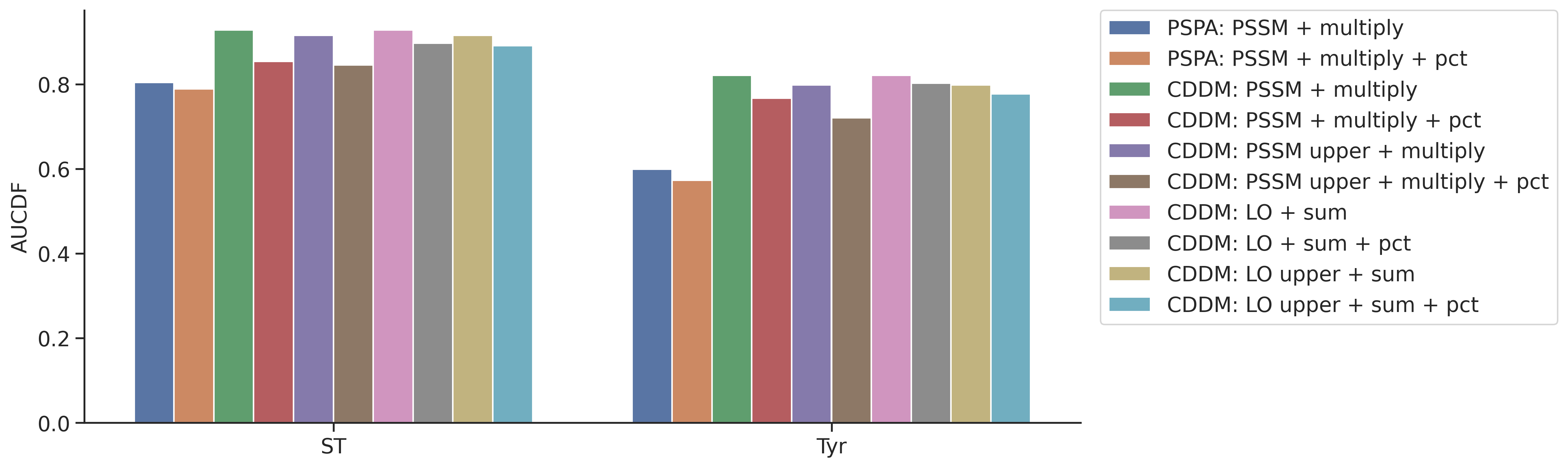
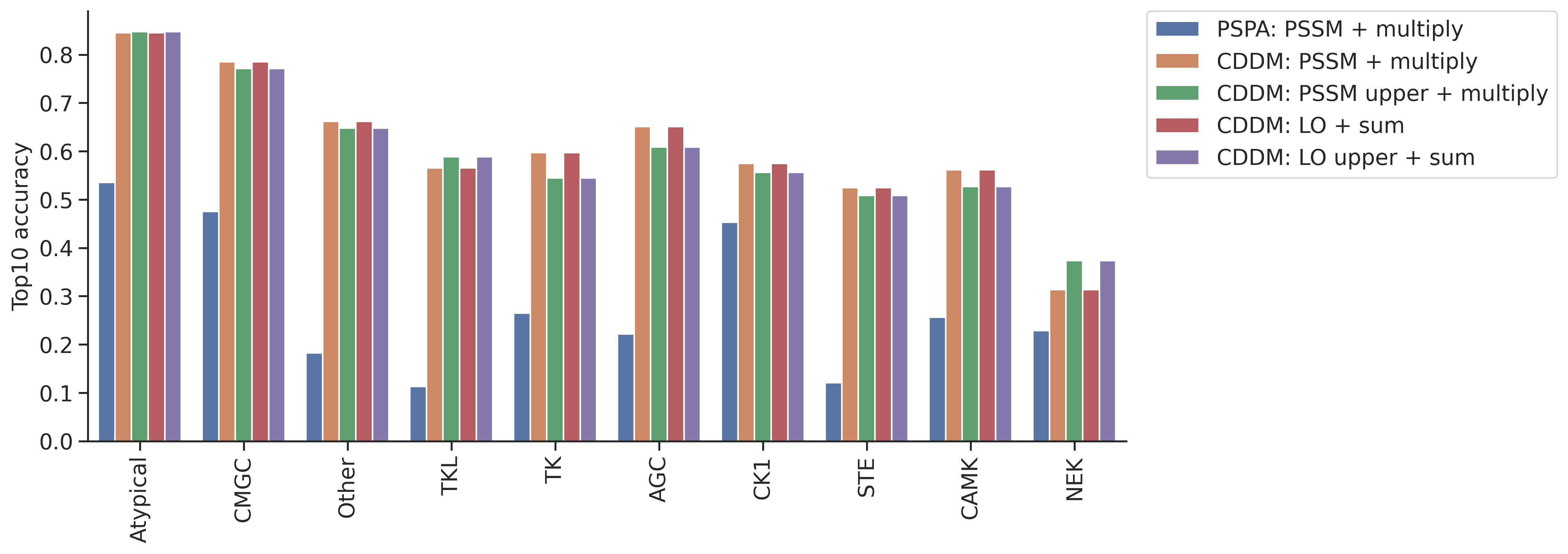
topk_df = pd.concat(topk_dfs,axis=1)topk_df.columns = ref_dict.keys()idx_ordered = topk_df.mean(1).sort_values(ascending=False).indextopk_df = topk_df.loc[idx_ordered]topk_df = topk_df.reset_index(names='group')plot_group_bar(topk_df,value_cols=topk_df.columns[1:],group='group')
plt.ylabel('Top10 accuracy')Text(0, 0.5, 'Top10 accuracy')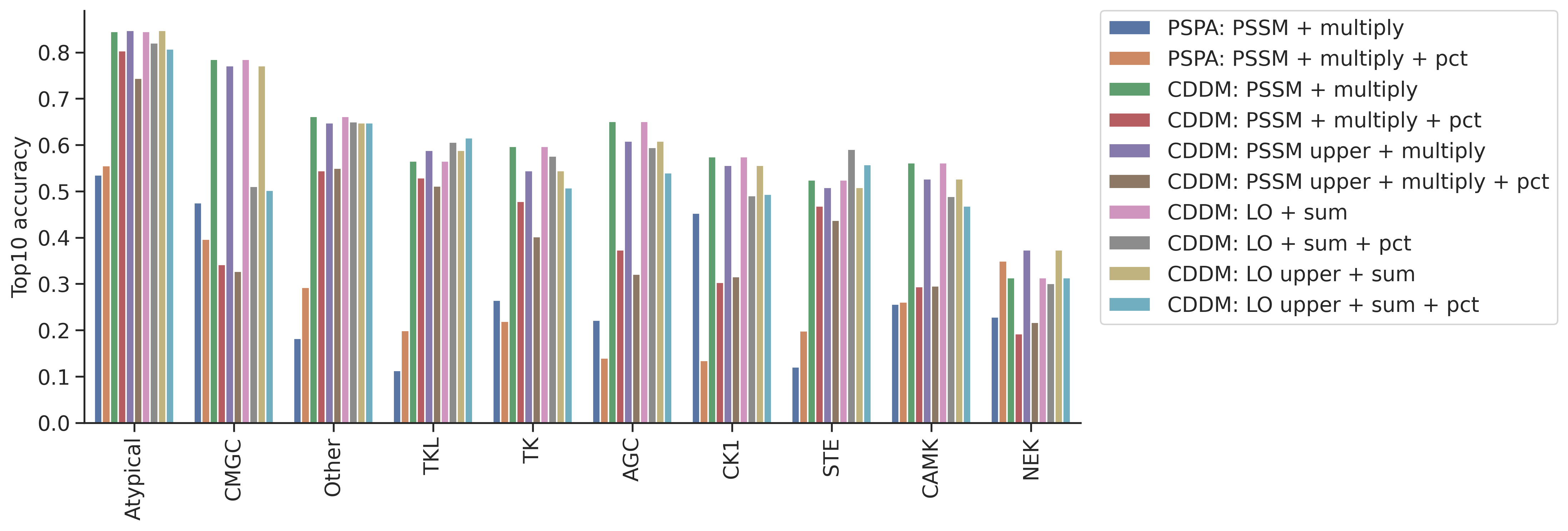
col = topk_df.columns[~topk_df.columns.str.contains('pct')][1:]plot_group_bar(topk_df,value_cols=col,group='group')
plt.ylabel('Top10 accuracy')
save_svg('img.svg')