# katlas
from katlas.core import *
from katlas.plot import *
# utils
import pandas as pd, numpy as np, seaborn as sns
from matplotlib import pyplot as plt
from functools import reduce, partial
from tqdm import tqdm
# statistics
from scipy.stats import ttest_rel,ttest_ind
from statsmodels.stats.multitest import multipletests
# disable warning
import warnings
warnings.filterwarnings("ignore", message="converting a masked element to nan")
pd.set_option('display.precision', 15)
set_sns()Kinase enrichment analysis - ATM
In this session, we will analyze the differential change of phosphorylation sites in phosphoproteomics dataset.
Setup
Data
df = pd.read_csv('raw/ATM_radiation.csv')df.head()| SITE_+/-7_AA | Log2Fold-Change | |
|---|---|---|
| 0 | SRSGEDEsQEDVLMD | 2.617689667 |
| 1 | QEAREVNsQEEEEEE | 4.383139333 |
| 2 | KLFDVCGsQDFESDL | 3.119948000 |
| 3 | ESEKQQDsQPEEVMD | 2.178596333 |
| 4 | QDGEVQLsQNDDKTK | 0.767963900 |
Substrate scoring
# pspa_out = predict_kinase_df(azd,seq_col='site_seq2', **param_PSPA)
cddm_out = predict_kinase_df(df,seq_col='SITE_+/-7_AA', **param_CDDM_upper)input dataframe has a length 37
Preprocessing
Finish preprocessing
Calculating position: [-7, -6, -5, -4, -3, -2, -1, 0, 1, 2, 3, 4, 5, 6, 7]100%|██████████| 289/289 [00:03<00:00, 84.33it/s]Kinase enrichment
def top_kinases(site_row,top_n=5):
# Sort the row in descending order and get the top n kinases
top_kinases = site_row.sort_values(ascending=False).head(top_n)
# Get the counts of the top kinases
kinase_counts = top_kinases.index.value_counts()
return kinase_countsfunc = partial(top_kinases,top_n=10)cnt_df = cddm_out.apply(func,axis=1)cnt = cnt_df.sum()w_cnt_df = cnt_df.multiply(df['Log2Fold-Change'],axis=0)w_cnt = w_cnt_df.sum()w_cntAKT1 4.160016533
AKT3 4.160016533
ALK2 15.193937901
ALK4 2.591647667
AMPKA1 3.844430200
...
TLK2 0.859286567
TNIK 0.859286567
TSSK1 9.668330334
TSSK2 13.435776167
ULK3 4.405065666
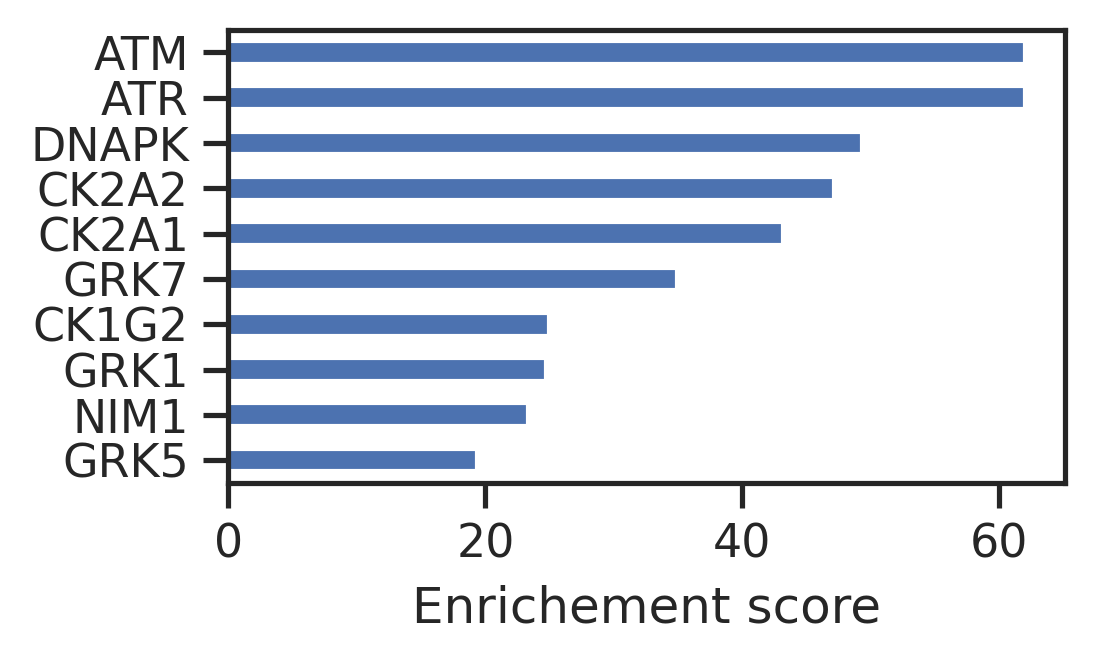
Length: 88, dtype: float64enrich = pd.concat([cnt,w_cnt],axis=1)enrich.columns = ['count','weighted_count']enrich = enrich.sort_values('weighted_count')set_sns()enrich.tail(10).weighted_count.plot.barh(figsize=(3.6,2))
plt.xlabel('Enrichement score');