from Bio.Seq import Seq
from Bio.SeqRecord import SeqRecord
from Bio import SeqIO, AlignIO
import pandas as pdAlignment
Prepare fasta file for clustalo
kd=pd.read_excel('out/uniprot_kd.xlsx')kd| kd_ID | Uniprot | Entry Name | Protein names | Gene Names | Gene Names (primary) | Organism | kd_note | kd_evidence | kd_start | ... | Reactome | ComplexPortal | Subcellular location [CC] | Gene Ontology (biological process) | Tissue specificity | Interacts with | Subunit structure | Function [CC] | Activity regulation | full_seq | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | A0A075F7E9_LERK1_ORYSI_KD1 | A0A075F7E9 | LERK1_ORYSI | G-type lectin S-receptor-like serine/threonine... | LECRK1 LECRK OsI_14840 | LECRK1 | Oryza sativa subsp. indica (Rice) | Protein kinase | ECO:0000255|PROSITE-ProRule:PRU00159 | 523 | ... | NaN | NaN | SUBCELLULAR LOCATION: Membrane {ECO:0000255}; ... | defense response [GO:0006952]; response to oth... | TISSUE SPECIFICITY: Expressed in plumules, rad... | NaN | SUBUNIT: Interacts (via kinase domain) with AD... | FUNCTION: Involved in innate immunity. Require... | NaN | MVALLLFPMLLQLLSPTCAQTQKNITLGSTLAPQGPASSWLSPSGD... |
| 1 | A0A078BQP2_GCY25_CAEEL_KD1 | A0A078BQP2 | GCY25_CAEEL | Receptor-type guanylate cyclase gcy-25 (EC 4.6... | gcy-25 Y105C5B.2 | gcy-25 | Caenorhabditis elegans | Protein kinase | ECO:0000255|PROSITE-ProRule:PRU00159 | 464 | ... | R-CEL-2514859; | NaN | SUBCELLULAR LOCATION: Cell membrane {ECO:00003... | cGMP biosynthetic process [GO:0006182]; intrac... | TISSUE SPECIFICITY: Expressed in AQR, PQR and ... | NaN | NaN | FUNCTION: Guanylate cyclase involved in the pr... | NaN | MLLLLLLLKISTFVDSFQIGHLEFENSNETRILEICMKNAGSWRDH... |
| 2 | A0A078CGE6_M3KE1_BRANA_KD1 | A0A078CGE6 | M3KE1_BRANA | MAP3K epsilon protein kinase 1 (BnM3KE1) (EC 2... | M3KE1 BnaA03g30290D GSBRNA2T00111755001 | M3KE1 | Brassica napus (Rape) | Protein kinase | ECO:0000255|PROSITE-ProRule:PRU00159 | 20 | ... | NaN | NaN | SUBCELLULAR LOCATION: Cytoplasm, cytoskeleton,... | cell division [GO:0051301]; protein autophosph... | TISSUE SPECIFICITY: Expressed in both the spor... | NaN | NaN | FUNCTION: Serine/threonine-protein kinase invo... | NaN | MARQMTSSQFHKSKTLDNKYMLGDEIGKGAYGRVYIGLDLENGDFV... |
| 3 | A0A0G2K344_PK3CA_RAT_KD1 | A0A0G2K344 | PK3CA_RAT | Phosphatidylinositol 4,5-bisphosphate 3-kinase... | Pik3ca | Pik3ca | Rattus norvegicus (Rat) | PI3K/PI4K catalytic | ECO:0000255|PROSITE-ProRule:PRU00269 | 765 | ... | R-RNO-109704;R-RNO-112399;R-RNO-114604;R-RNO-1... | NaN | NaN | actin cytoskeleton organization [GO:0030036]; ... | TISSUE SPECIFICITY: Detected in the hypothalam... | NaN | SUBUNIT: Heterodimer of a catalytic subunit PI... | FUNCTION: Phosphoinositide-3-kinase (PI3K) pho... | NaN | MPPRPSSGELWGIHLMPPRILVECLLPNGMIVTLECLREATLVTIK... |
| 4 | A0A0H2ZM62_HK06_STRP2_KD1 | A0A0H2ZM62 | HK06_STRP2 | Sensor histidine protein kinase HK06 (EC 2.7.1... | hk06 SPD_2019 | hk06 | Streptococcus pneumoniae serotype 2 (strain D3... | Histidine kinase | ECO:0000255|PROSITE-ProRule:PRU00107 | 239 | ... | NaN | NaN | SUBCELLULAR LOCATION: Cell membrane {ECO:00003... | NaN | NaN | NaN | NaN | FUNCTION: Member of the two-component regulato... | NaN | MIKNPKLLTKSFLRSFAILGGVGLVIHIAIYLTFPFYYIQLEGEKF... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 5531 | W0LYS5_CAMKI_MACNP_KD1 | W0LYS5 | CAMKI_MACNP | Calcium/calmodulin-dependent protein kinase ty... | CaMKI | CaMKI | Macrobrachium nipponense (Oriental river shrim... | Protein kinase | ECO:0000255|PROSITE-ProRule:PRU00159 | 31 | ... | NaN | NaN | NaN | molting cycle process [GO:0022404] | TISSUE SPECIFICITY: Highly expressed in hepato... | NaN | NaN | FUNCTION: Calcium/calmodulin-dependent protein... | ACTIVITY REGULATION: Activated by Ca(2+)/calmo... | MPLFGSKKETAKKSSKKDKDEGKMPAVEDKYILKDLLGTGAFSQVR... |
| 5532 | W0T9X4_ATG1_KLUMD_KD1 | W0T9X4 | ATG1_KLUMD | Serine/threonine-protein kinase ATG1 (EC 2.7.1... | ATG1 KLMA_30321 | ATG1 | Kluyveromyces marxianus (strain DMKU3-1042 / B... | Protein kinase | ECO:0000255|PROSITE-ProRule:PRU00159 | 21 | ... | NaN | NaN | SUBCELLULAR LOCATION: Cytoplasm {ECO:0000250|U... | autophagosome assembly [GO:0000045]; autophagy... | NaN | W0TA43 | SUBUNIT: Homodimer (By similarity). Dimerizati... | FUNCTION: Serine/threonine protein kinase invo... | NaN | MSSESHGKAVAKAIRLPTENYTVEKEIGKGSFAIVYKGVSLRDGRN... |
| 5533 | W7JX98_KGP_PLAFO_KD1 | W7JX98 | KGP_PLAFO | cGMP-dependent protein kinase (EC 2.7.11.12) | PKG PFNF54_05395 | PKG | Plasmodium falciparum (isolate NF54) | Protein kinase | ECO:0000255|PROSITE-ProRule:PRU00159 | 541 | ... | NaN | NaN | SUBCELLULAR LOCATION: Cytoplasm {ECO:0000250|U... | protein phosphorylation [GO:0006468] | NaN | NaN | SUBUNIT: Monomer. {ECO:0000250|UniProtKB:Q8I719}. | FUNCTION: Serine/threonine protein kinase whic... | ACTIVITY REGULATION: Activated by cGMP. Not ac... | MEEDDNLKKGNERNKKKAIFSNDDFTGEDSLMEDHLELREKLSEDI... |
| 5534 | X5M5N0_WNK_CAEEL_KD1 | X5M5N0 | WNK_CAEEL | Serine/threonine-protein kinase WNK (EC 2.7.11... | wnk-1 C46C2.1 | wnk-1 | Caenorhabditis elegans | Protein kinase | ECO:0000255|PROSITE-ProRule:PRU00159 | 334 | ... | NaN | NaN | SUBCELLULAR LOCATION: Cytoplasm {ECO:0000250|U... | cell volume homeostasis [GO:0006884]; cellular... | TISSUE SPECIFICITY: Expressed in pharynx, nerv... | G5EEN4 | SUBUNIT: Interacts with gck-3 (via C-terminus)... | FUNCTION: Serine/threonine-protein kinase comp... | ACTIVITY REGULATION: Activated in response to ... | MPDSITNGGRPPAPPSSVSSTTASTTGNFGTRRRLVNRIKKVDELH... |
| 5535 | X5M8U1_GCY17_CAEEL_KD1 | X5M8U1 | GCY17_CAEEL | Receptor-type guanylate cyclase gcy-17 (EC 4.6... | gcy-17 W03F11.2 | gcy-17 | Caenorhabditis elegans | Protein kinase | ECO:0000255|PROSITE-ProRule:PRU00159 | 535 | ... | R-CEL-2514859; | NaN | SUBCELLULAR LOCATION: Cell membrane {ECO:00003... | cGMP biosynthetic process [GO:0006182]; intrac... | TISSUE SPECIFICITY: Expressed in PHA sensory n... | NaN | NaN | FUNCTION: Guanylate cyclase involved in the pr... | NaN | MLFLRLFIFTPFLILANCQARRTIKVGLLFVQNVSSLQVGIGYRTS... |
5536 rows × 27 columns
def get_fasta(df,seq_col='kd_seq',id_col='kd_ID',path='out.fasta'):
"Generate fasta file from sequences."
records = [
SeqRecord(Seq(row[seq_col]), id=row[id_col], description="")
for _, row in df.iterrows()
]
SeqIO.write(records, path, "fasta")
print(len(records))get_fasta(kd,path='raw/kinase_domains.fasta')5536For human only:
human = kd[kd.Organism=='Homo sapiens (Human)']get_fasta(human,path="raw/human_kinase_domains.fasta")539for PI3K catalytic domain
pi3k = kd[kd.kd_note=='PI3K/PI4K catalytic']get_fasta(pi3k,path="raw/pi3k_kinase_domains.fasta")168Run clustalo
sudo apt-get update
sudo apt-get install clustalo
clustalo -i kinase_domains.fasta -o kinase_domains.aln --force --outfmt=clu# import subprocess
# subprocess.run([
# "clustalo", "-i", "kinase_domains.fasta",
# "-o", "kinase_domains.aln", "--force", "--outfmt=clu"
# ])Analyze alignment
alignment = AlignIO.read("raw/kinase_domains.aln", "clustal")Turns in to dataframe:
alignment_array = [list(str(record.seq)) for record in alignment]
df = pd.DataFrame(alignment_array)df.columns = df.columns+1df.index=kd.kd_ID# df.to_parquet('output/uniprot_kd_align.parquet')df.head()| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ... | 3425 | 3426 | 3427 | 3428 | 3429 | 3430 | 3431 | 3432 | 3433 | 3434 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kd_ID | |||||||||||||||||||||
| A0A075F7E9_LERK1_ORYSI_KD1 | - | - | - | - | - | - | - | - | - | - | ... | - | - | - | - | - | - | - | - | - | - |
| A0A078BQP2_GCY25_CAEEL_KD1 | - | - | - | - | - | - | - | - | - | - | ... | - | - | - | - | - | - | - | - | - | - |
| A0A078CGE6_M3KE1_BRANA_KD1 | - | - | - | - | - | - | - | - | - | - | ... | - | - | - | - | - | - | - | - | - | - |
| A0A0G2K344_PK3CA_RAT_KD1 | - | - | - | - | - | - | - | - | - | - | ... | - | - | - | - | - | - | - | - | - | - |
| A0A0H2ZM62_HK06_STRP2_KD1 | - | - | - | - | - | - | - | - | - | - | ... | - | - | - | - | - | - | - | - | - | - |
5 rows × 3434 columns
# Compute residue frequency at each position
counts_df = df.apply(lambda col: col.value_counts(), axis=0).fillna(0)
freq_df = counts_df.div(counts_df.sum(axis=0), axis=1)freq_df.head()| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ... | 3425 | 3426 | 3427 | 3428 | 3429 | 3430 | 3431 | 3432 | 3433 | 3434 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | 0.999819 | 0.999819 | 0.999819 | 0.999819 | 0.999819 | 0.999819 | 0.999819 | 0.999819 | 0.999819 | 0.999819 | ... | 0.999819 | 0.999819 | 0.999819 | 0.999819 | 0.999819 | 0.999819 | 0.999819 | 0.999819 | 0.999819 | 0.999819 |
| A | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| C | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| D | 0.000000 | 0.000181 | 0.000000 | 0.000000 | 0.000181 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000181 | 0.000181 | 0.000000 | 0.000000 | 0.000000 | 0.000181 | 0.000000 | 0.000000 | 0.000000 |
| E | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000181 | 0.000000 | 0.000000 |
5 rows × 3434 columns
# remove '-' first line
max_series=freq_df.iloc[1:,:]freq_max = pd.concat([max_series.idxmax(),max_series.max()],axis=1)freq_max.columns = ['aa','max_value']freq_max = freq_max.sort_values('max_value',ascending=False).reset_index(names='position')out = freq_max[freq_max.max_value>0.1]# out.to_csv('out/align_freq_max_aa.csv',index=False)out| position | aa | max_value | |
|---|---|---|---|
| 0 | 1549 | N | 0.815390 |
| 1 | 2618 | D | 0.809429 |
| 2 | 1724 | D | 0.800759 |
| 3 | 1525 | D | 0.791004 |
| 4 | 1730 | G | 0.775470 |
| ... | ... | ... | ... |
| 214 | 193 | E | 0.101879 |
| 215 | 640 | G | 0.101879 |
| 216 | 922 | L | 0.101337 |
| 217 | 603 | R | 0.101156 |
| 218 | 2581 | G | 0.100614 |
219 rows × 3 columns
Locate active motif & save
- 1549 N is after HRD motif
- 2618 D is around D[IV]WS motif
- 1724 D is DFG motif
- 1525 D is HRD motif
kd['D1']=(df[1525]=='D').astype(int) # HRDkd['D2'] = (df[1724]=='D').astype(int) #DFGkd['D3']=(df[2618]=='D').astype(int)kd['N1'] = (df[1549]=='N').astype(int)active_col = ['D1','D2']kd['active_D1_D2'] = (kd[active_col].sum(1)==len(active_col)).astype(int)active_all = kd[kd.active_D1_D2==1]active_all.shape(4209, 32)# active_all.to_excel('out/uniprot_kd_active_D1_D2.xlsx',index=False)# kd.to_excel('out/uniprot_kd_motif_labeled.xlsx',index=False)Take a look of their identity:
kd[kd.active_D1_D2==1].kd_note.value_counts()kd_note
Protein kinase 4005
PI3K/PI4K catalytic 71
Protein kinase 2 61
Protein kinase 1 37
Histidine kinase 34
Alpha-type protein kinase 1
Name: count, dtype: int64kd[kd.active_D1_D2==0].kd_note.value_counts()kd_note
Histidine kinase 591
Protein kinase 525
PI3K/PI4K catalytic 97
Protein kinase 1 36
Alpha-type protein kinase 21
Protein kinase; inactive 13
Protein kinase 2 12
HWE histidine kinase domain 11
Guanylate kinase-like 5
Histidine kinase 2 4
Histidine kinase 1 4
Amino-acid kinase domain (AAK) 3
Kinase domain 2
Histidine kinase; first part 1
Histidine kinase; second part 1
Protein kinase; truncated 1
Name: count, dtype: int64Merge with human PSPA
human = kd[kd.Organism=='Homo sapiens (Human)']human.active_D1_D2.value_counts()active_D1_D2
1 444
0 95
Name: count, dtype: int64human_active = human[human.active_D1_D2==1]human_active[human_active.Uniprot.duplicated(keep=False)]| kd_ID | Uniprot | Entry Name | Protein names | Gene Names | Gene Names (primary) | Organism | kd_note | kd_evidence | kd_start | ... | Interacts with | Subunit structure | Function [CC] | Activity regulation | full_seq | D1 | D2 | D3 | N1 | active_D1_D2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 988 | O75582_KS6A5_HUMAN_KD1 | O75582 | KS6A5_HUMAN | Ribosomal protein S6 kinase alpha-5 (S6K-alpha... | RPS6KA5 MSK1 | RPS6KA5 | Homo sapiens (Human) | Protein kinase 1 | ECO:0000255|PROSITE-ProRule:PRU00159 | 49 | ... | P67870; Q9UI47-2; Q9Y4C1; Q9NYL2; Q16539; Q9Y4C1 | SUBUNIT: Forms a complex with either MAPK1/ERK... | FUNCTION: Serine/threonine-protein kinase that... | ACTIVITY REGULATION: Activated by phosphorylat... | MEEEGGSSGGAAGTSADGGDGGEQLLTVKHELRTANLTGHAEKVGI... | 1 | 1 | 1 | 1 | 1 |
| 989 | O75582_KS6A5_HUMAN_KD2 | O75582 | KS6A5_HUMAN | Ribosomal protein S6 kinase alpha-5 (S6K-alpha... | RPS6KA5 MSK1 | RPS6KA5 | Homo sapiens (Human) | Protein kinase 2 | ECO:0000255|PROSITE-ProRule:PRU00159 | 426 | ... | P67870; Q9UI47-2; Q9Y4C1; Q9NYL2; Q16539; Q9Y4C1 | SUBUNIT: Forms a complex with either MAPK1/ERK... | FUNCTION: Serine/threonine-protein kinase that... | ACTIVITY REGULATION: Activated by phosphorylat... | MEEEGGSSGGAAGTSADGGDGGEQLLTVKHELRTANLTGHAEKVGI... | 1 | 1 | 1 | 1 | 1 |
| 990 | O75676_KS6A4_HUMAN_KD1 | O75676 | KS6A4_HUMAN | Ribosomal protein S6 kinase alpha-4 (S6K-alpha... | RPS6KA4 MSK2 | RPS6KA4 | Homo sapiens (Human) | Protein kinase 1 | ECO:0000255|PROSITE-ProRule:PRU00159 | 33 | ... | Q16539; O14901 | SUBUNIT: Forms a complex with either MAPK1/ERK... | FUNCTION: Serine/threonine-protein kinase that... | ACTIVITY REGULATION: Activated by phosphorylat... | MGDEDDDESCAVELRITEANLTGHEEKVSVENFELLKVLGTGAYGK... | 1 | 1 | 1 | 1 | 1 |
| 991 | O75676_KS6A4_HUMAN_KD2 | O75676 | KS6A4_HUMAN | Ribosomal protein S6 kinase alpha-4 (S6K-alpha... | RPS6KA4 MSK2 | RPS6KA4 | Homo sapiens (Human) | Protein kinase 2 | ECO:0000255|PROSITE-ProRule:PRU00159 | 411 | ... | Q16539; O14901 | SUBUNIT: Forms a complex with either MAPK1/ERK... | FUNCTION: Serine/threonine-protein kinase that... | ACTIVITY REGULATION: Activated by phosphorylat... | MGDEDDDESCAVELRITEANLTGHEEKVSVENFELLKVLGTGAYGK... | 1 | 1 | 1 | 1 | 1 |
| 1918 | P51812_KS6A3_HUMAN_KD1 | P51812 | KS6A3_HUMAN | Ribosomal protein S6 kinase alpha-3 (S6K-alpha... | RPS6KA3 ISPK1 MAPKAPK1B RSK2 | RPS6KA3 | Homo sapiens (Human) | Protein kinase 1 | ECO:0000255|PROSITE-ProRule:PRU00159 | 68 | ... | P46379-2; P67870; P09471; P08238; O14901; P284... | SUBUNIT: Forms a complex with either MAPK1/ERK... | FUNCTION: Serine/threonine-protein kinase that... | ACTIVITY REGULATION: Upon extracellular signal... | MPLAQLADPWQKMAVESPSDSAENGQQIMDEPMGEEEINPQTEEVS... | 1 | 1 | 1 | 1 | 1 |
| 1919 | P51812_KS6A3_HUMAN_KD2 | P51812 | KS6A3_HUMAN | Ribosomal protein S6 kinase alpha-3 (S6K-alpha... | RPS6KA3 ISPK1 MAPKAPK1B RSK2 | RPS6KA3 | Homo sapiens (Human) | Protein kinase 2 | ECO:0000255|PROSITE-ProRule:PRU00159 | 422 | ... | P46379-2; P67870; P09471; P08238; O14901; P284... | SUBUNIT: Forms a complex with either MAPK1/ERK... | FUNCTION: Serine/threonine-protein kinase that... | ACTIVITY REGULATION: Upon extracellular signal... | MPLAQLADPWQKMAVESPSDSAENGQQIMDEPMGEEEINPQTEEVS... | 1 | 1 | 1 | 1 | 1 |
| 2560 | Q15349_KS6A2_HUMAN_KD1 | Q15349 | KS6A2_HUMAN | Ribosomal protein S6 kinase alpha-2 (S6K-alpha... | RPS6KA2 MAPKAPK1C RSK3 | RPS6KA2 | Homo sapiens (Human) | Protein kinase 1 | ECO:0000255|PROSITE-ProRule:PRU00159 | 59 | ... | P05067; P15056; P67870; O14901; P28482; Q02156... | SUBUNIT: Forms a complex with either MAPK1/ERK... | FUNCTION: Serine/threonine-protein kinase that... | ACTIVITY REGULATION: Upon extracellular signal... | MDLSMKKFAVRRFFSVYLRRKSRSKSSSLSRLEEEGVVKEIDISHH... | 1 | 1 | 1 | 1 | 1 |
| 2561 | Q15349_KS6A2_HUMAN_KD2 | Q15349 | KS6A2_HUMAN | Ribosomal protein S6 kinase alpha-2 (S6K-alpha... | RPS6KA2 MAPKAPK1C RSK3 | RPS6KA2 | Homo sapiens (Human) | Protein kinase 2 | ECO:0000255|PROSITE-ProRule:PRU00159 | 415 | ... | P05067; P15056; P67870; O14901; P28482; Q02156... | SUBUNIT: Forms a complex with either MAPK1/ERK... | FUNCTION: Serine/threonine-protein kinase that... | ACTIVITY REGULATION: Upon extracellular signal... | MDLSMKKFAVRRFFSVYLRRKSRSKSSSLSRLEEEGVVKEIDISHH... | 1 | 1 | 1 | 1 | 1 |
| 2563 | Q15418_KS6A1_HUMAN_KD1 | Q15418 | KS6A1_HUMAN | Ribosomal protein S6 kinase alpha-1 (S6K-alpha... | RPS6KA1 MAPKAPK1A RSK1 | RPS6KA1 | Homo sapiens (Human) | Protein kinase 1 | ECO:0000255|PROSITE-ProRule:PRU00159 | 62 | ... | O43823; Q16543; P46527; P08238; P28482; P04271... | SUBUNIT: Forms a complex with either MAPK1/ERK... | FUNCTION: Serine/threonine-protein kinase that... | ACTIVITY REGULATION: Upon extracellular signal... | MPLAQLKEPWPLMELVPLDPENGQTSGEEAGLQPSKDEGVLKEISI... | 1 | 1 | 1 | 1 | 1 |
| 2564 | Q15418_KS6A1_HUMAN_KD2 | Q15418 | KS6A1_HUMAN | Ribosomal protein S6 kinase alpha-1 (S6K-alpha... | RPS6KA1 MAPKAPK1A RSK1 | RPS6KA1 | Homo sapiens (Human) | Protein kinase 2 | ECO:0000255|PROSITE-ProRule:PRU00159 | 418 | ... | O43823; Q16543; P46527; P08238; P28482; P04271... | SUBUNIT: Forms a complex with either MAPK1/ERK... | FUNCTION: Serine/threonine-protein kinase that... | ACTIVITY REGULATION: Upon extracellular signal... | MPLAQLKEPWPLMELVPLDPENGQTSGEEAGLQPSKDEGVLKEISI... | 1 | 1 | 1 | 1 | 1 |
| 2568 | Q15772_SPEG_HUMAN_KD1 | Q15772 | SPEG_HUMAN | Striated muscle preferentially expressed prote... | SPEG APEG1 KIAA1297 | SPEG | Homo sapiens (Human) | Protein kinase 1 | ECO:0000255|PROSITE-ProRule:PRU00159 | 1601 | ... | Q99873; O75031; Q6FHY5; Q9NRD5; Q99873-3 | SUBUNIT: Interacts with MTM1. Isoform 3 is fou... | FUNCTION: Isoform 3 may have a role in regulat... | NaN | MQKARGTRGEDAGTRAPPSPGVPPKRAKVGAGGGAPVAVAGAPVFL... | 1 | 1 | 1 | 1 | 1 |
| 2569 | Q15772_SPEG_HUMAN_KD2 | Q15772 | SPEG_HUMAN | Striated muscle preferentially expressed prote... | SPEG APEG1 KIAA1297 | SPEG | Homo sapiens (Human) | Protein kinase 2 | ECO:0000255|PROSITE-ProRule:PRU00159 | 2966 | ... | Q99873; O75031; Q6FHY5; Q9NRD5; Q99873-3 | SUBUNIT: Interacts with MTM1. Isoform 3 is fou... | FUNCTION: Isoform 3 may have a role in regulat... | NaN | MQKARGTRGEDAGTRAPPSPGVPPKRAKVGAGGGAPVAVAGAPVFL... | 1 | 1 | 1 | 1 | 1 |
| 3480 | Q5VST9_OBSCN_HUMAN_KD1 | Q5VST9 | OBSCN_HUMAN | Obscurin (EC 2.7.11.1) (Obscurin-RhoGEF) (Obsc... | OBSCN KIAA1556 KIAA1639 | OBSCN | Homo sapiens (Human) | Protein kinase 1 | ECO:0000255|PROSITE-ProRule:PRU00159 | 6468 | ... | Q8WZ42; P16157; P16157-17 | SUBUNIT: Interacts (via protein kinase domain ... | FUNCTION: Structural component of striated mus... | NaN | MDQPQFSGAPRFLTRPKAFVVSVGKDATLSCQIVGNPTPQVSWEKD... | 1 | 1 | 1 | 1 | 1 |
| 3481 | Q5VST9_OBSCN_HUMAN_KD2 | Q5VST9 | OBSCN_HUMAN | Obscurin (EC 2.7.11.1) (Obscurin-RhoGEF) (Obsc... | OBSCN KIAA1556 KIAA1639 | OBSCN | Homo sapiens (Human) | Protein kinase 2 | ECO:0000255|PROSITE-ProRule:PRU00159 | 7672 | ... | Q8WZ42; P16157; P16157-17 | SUBUNIT: Interacts (via protein kinase domain ... | FUNCTION: Structural component of striated mus... | NaN | MDQPQFSGAPRFLTRPKAFVVSVGKDATLSCQIVGNPTPQVSWEKD... | 1 | 1 | 1 | 1 | 1 |
| 5338 | Q9UK32_KS6A6_HUMAN_KD1 | Q9UK32 | KS6A6_HUMAN | Ribosomal protein S6 kinase alpha-6 (S6K-alpha... | RPS6KA6 RSK4 | RPS6KA6 | Homo sapiens (Human) | Protein kinase 1 | ECO:0000255|PROSITE-ProRule:PRU00159 | 73 | ... | Q7Z698 | SUBUNIT: Forms a complex with MAPK3/ERK1 but n... | FUNCTION: Constitutively active serine/threoni... | ACTIVITY REGULATION: Constitutively activated ... | MLPFAPQDEPWDREMEVFSGGGASSGEVNGLKMVDEPMEEGEADSC... | 1 | 1 | 1 | 1 | 1 |
| 5339 | Q9UK32_KS6A6_HUMAN_KD2 | Q9UK32 | KS6A6_HUMAN | Ribosomal protein S6 kinase alpha-6 (S6K-alpha... | RPS6KA6 RSK4 | RPS6KA6 | Homo sapiens (Human) | Protein kinase 2 | ECO:0000255|PROSITE-ProRule:PRU00159 | 426 | ... | Q7Z698 | SUBUNIT: Forms a complex with MAPK3/ERK1 but n... | FUNCTION: Constitutively active serine/threoni... | ACTIVITY REGULATION: Constitutively activated ... | MLPFAPQDEPWDREMEVFSGGGASSGEVNGLKMVDEPMEEGEADSC... | 1 | 1 | 1 | 1 | 1 |
16 rows × 32 columns
from katlas.data import *info=Data.get_kinase_info()kinase_map = info[['kinase','uniprot']]kinase_map = kinase_map[~kinase_map.kinase.str.contains('_b')]kinase_map = kinase_map.set_index('kinase')['uniprot']kinase_map = kinase_map['uniprot']kinase_map = kinase_map.drop_duplicates()# kinase_map.to_csv('out/kinase_map.csv')kinase_mapkinase
AAK1 Q2M2I8
ABL1 P00519
ABL2 P42684
TNK2 Q07912
ACVR2A P27037
...
YSK1 O00506
ZAK Q9NYL2
ZAP70 P43403
EEF2K O00418
FAM20C Q8IXL6
Name: uniprot, Length: 509, dtype: objectpspa = Data.get_pspa_all_norm()pspa.index.duplicated().sum()0idx = pspa.index.str.split('_').str[0]idx.map(kinase_map).isna().sum()0pspa = pspa.reset_index()uniprot = pd.DataFrame(idx.map(kinase_map))uniprot.columns=['uniprot']pspa = pd.concat([uniprot,pspa],axis=1)# pspa.to_csv('out/pspa_uniprot.csv',index=False)PSPA with duplicated uniprot:
pspa[pspa.uniprot.duplicated(keep=False)].sort_values('uniprot')| uniprot | kinase | -5P | -5G | -5A | -5C | -5S | -5T | -5V | -5I | ... | 5H | 5K | 5R | 5Q | 5N | 5D | 5E | 5s | 5t | 5y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 22 | Q13873 | BMPR2 | 0.0558 | 0.0621 | 0.0638 | 0.0716 | 0.0571 | 0.0571 | 0.0597 | 0.0571 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 309 | Q13873 | BMPR2_TYR | 0.0580 | 0.0617 | 0.0603 | 0.0661 | 0.0613 | 0.0613 | 0.0566 | 0.0580 | ... | 0.0660 | 0.0665 | 0.0766 | 0.0607 | 0.0619 | 0.0648 | 0.0649 | 0.0702 | 0.0702 | 0.0653 |
| 212 | Q15118 | PDHK1 | 0.0451 | 0.0697 | 0.0594 | 0.0625 | 0.0594 | 0.0594 | 0.0573 | 0.0590 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 369 | Q15118 | PDHK1_TYR | 0.0590 | 0.0608 | 0.0528 | 0.0620 | 0.0608 | 0.0608 | 0.0439 | 0.0528 | ... | 0.0676 | 0.0490 | 0.0539 | 0.0620 | 0.0647 | 0.0949 | 0.0797 | 0.0633 | 0.0633 | 0.0820 |
| 213 | Q16654 | PDHK4 | 0.0452 | 0.0645 | 0.0665 | 0.0672 | 0.0622 | 0.0622 | 0.0515 | 0.0619 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 371 | Q16654 | PDHK4_TYR | 0.0697 | 0.0690 | 0.0545 | 0.0772 | 0.0642 | 0.0642 | 0.0426 | 0.0483 | ... | 0.0642 | 0.0486 | 0.0498 | 0.0755 | 0.0661 | 0.0836 | 0.0830 | 0.0686 | 0.0686 | 0.0892 |
| 221 | Q9BXM7 | PINK1 | 0.0516 | 0.0474 | 0.0612 | 0.0570 | 0.0523 | 0.0523 | 0.0474 | 0.0507 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 372 | Q9BXM7 | PINK1_TYR | 0.0569 | 0.0585 | 0.0572 | 0.0643 | 0.0579 | 0.0579 | 0.0546 | 0.0555 | ... | 0.0666 | 0.0818 | 0.1049 | 0.0636 | 0.0635 | 0.0525 | 0.0508 | 0.0516 | 0.0516 | 0.0611 |
8 rows × 238 columns
pspa_unique_uniprot = pspa[~pspa.uniprot.duplicated()]Also remove other TYR due to their overall low specificity
pspa_no_TYR = pspa_unique_uniprot[~pspa_unique_uniprot.kinase.str.contains('_TYR')]pspa_no_TYR = pspa_no_TYR.dropna(axis=1)# pspa_no_TYR.to_csv('out/pspa_uniprot_unique_no_TYR.csv',index=False)active_kd = pd.read_excel('out/uniprot_kd_active_D1_D2.xlsx')human_active[human_active.Uniprot.duplicated()]| kd_ID | Uniprot | Entry Name | Protein names | Gene Names | Gene Names (primary) | Organism | kd_note | kd_evidence | kd_start | ... | Interacts with | Subunit structure | Function [CC] | Activity regulation | full_seq | D1 | D2 | D3 | N1 | active_D1_D2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 989 | O75582_KS6A5_HUMAN_KD2 | O75582 | KS6A5_HUMAN | Ribosomal protein S6 kinase alpha-5 (S6K-alpha... | RPS6KA5 MSK1 | RPS6KA5 | Homo sapiens (Human) | Protein kinase 2 | ECO:0000255|PROSITE-ProRule:PRU00159 | 426 | ... | P67870; Q9UI47-2; Q9Y4C1; Q9NYL2; Q16539; Q9Y4C1 | SUBUNIT: Forms a complex with either MAPK1/ERK... | FUNCTION: Serine/threonine-protein kinase that... | ACTIVITY REGULATION: Activated by phosphorylat... | MEEEGGSSGGAAGTSADGGDGGEQLLTVKHELRTANLTGHAEKVGI... | 1 | 1 | 1 | 1 | 1 |
| 991 | O75676_KS6A4_HUMAN_KD2 | O75676 | KS6A4_HUMAN | Ribosomal protein S6 kinase alpha-4 (S6K-alpha... | RPS6KA4 MSK2 | RPS6KA4 | Homo sapiens (Human) | Protein kinase 2 | ECO:0000255|PROSITE-ProRule:PRU00159 | 411 | ... | Q16539; O14901 | SUBUNIT: Forms a complex with either MAPK1/ERK... | FUNCTION: Serine/threonine-protein kinase that... | ACTIVITY REGULATION: Activated by phosphorylat... | MGDEDDDESCAVELRITEANLTGHEEKVSVENFELLKVLGTGAYGK... | 1 | 1 | 1 | 1 | 1 |
| 1919 | P51812_KS6A3_HUMAN_KD2 | P51812 | KS6A3_HUMAN | Ribosomal protein S6 kinase alpha-3 (S6K-alpha... | RPS6KA3 ISPK1 MAPKAPK1B RSK2 | RPS6KA3 | Homo sapiens (Human) | Protein kinase 2 | ECO:0000255|PROSITE-ProRule:PRU00159 | 422 | ... | P46379-2; P67870; P09471; P08238; O14901; P284... | SUBUNIT: Forms a complex with either MAPK1/ERK... | FUNCTION: Serine/threonine-protein kinase that... | ACTIVITY REGULATION: Upon extracellular signal... | MPLAQLADPWQKMAVESPSDSAENGQQIMDEPMGEEEINPQTEEVS... | 1 | 1 | 1 | 1 | 1 |
| 2561 | Q15349_KS6A2_HUMAN_KD2 | Q15349 | KS6A2_HUMAN | Ribosomal protein S6 kinase alpha-2 (S6K-alpha... | RPS6KA2 MAPKAPK1C RSK3 | RPS6KA2 | Homo sapiens (Human) | Protein kinase 2 | ECO:0000255|PROSITE-ProRule:PRU00159 | 415 | ... | P05067; P15056; P67870; O14901; P28482; Q02156... | SUBUNIT: Forms a complex with either MAPK1/ERK... | FUNCTION: Serine/threonine-protein kinase that... | ACTIVITY REGULATION: Upon extracellular signal... | MDLSMKKFAVRRFFSVYLRRKSRSKSSSLSRLEEEGVVKEIDISHH... | 1 | 1 | 1 | 1 | 1 |
| 2564 | Q15418_KS6A1_HUMAN_KD2 | Q15418 | KS6A1_HUMAN | Ribosomal protein S6 kinase alpha-1 (S6K-alpha... | RPS6KA1 MAPKAPK1A RSK1 | RPS6KA1 | Homo sapiens (Human) | Protein kinase 2 | ECO:0000255|PROSITE-ProRule:PRU00159 | 418 | ... | O43823; Q16543; P46527; P08238; P28482; P04271... | SUBUNIT: Forms a complex with either MAPK1/ERK... | FUNCTION: Serine/threonine-protein kinase that... | ACTIVITY REGULATION: Upon extracellular signal... | MPLAQLKEPWPLMELVPLDPENGQTSGEEAGLQPSKDEGVLKEISI... | 1 | 1 | 1 | 1 | 1 |
| 2569 | Q15772_SPEG_HUMAN_KD2 | Q15772 | SPEG_HUMAN | Striated muscle preferentially expressed prote... | SPEG APEG1 KIAA1297 | SPEG | Homo sapiens (Human) | Protein kinase 2 | ECO:0000255|PROSITE-ProRule:PRU00159 | 2966 | ... | Q99873; O75031; Q6FHY5; Q9NRD5; Q99873-3 | SUBUNIT: Interacts with MTM1. Isoform 3 is fou... | FUNCTION: Isoform 3 may have a role in regulat... | NaN | MQKARGTRGEDAGTRAPPSPGVPPKRAKVGAGGGAPVAVAGAPVFL... | 1 | 1 | 1 | 1 | 1 |
| 3481 | Q5VST9_OBSCN_HUMAN_KD2 | Q5VST9 | OBSCN_HUMAN | Obscurin (EC 2.7.11.1) (Obscurin-RhoGEF) (Obsc... | OBSCN KIAA1556 KIAA1639 | OBSCN | Homo sapiens (Human) | Protein kinase 2 | ECO:0000255|PROSITE-ProRule:PRU00159 | 7672 | ... | Q8WZ42; P16157; P16157-17 | SUBUNIT: Interacts (via protein kinase domain ... | FUNCTION: Structural component of striated mus... | NaN | MDQPQFSGAPRFLTRPKAFVVSVGKDATLSCQIVGNPTPQVSWEKD... | 1 | 1 | 1 | 1 | 1 |
| 5339 | Q9UK32_KS6A6_HUMAN_KD2 | Q9UK32 | KS6A6_HUMAN | Ribosomal protein S6 kinase alpha-6 (S6K-alpha... | RPS6KA6 RSK4 | RPS6KA6 | Homo sapiens (Human) | Protein kinase 2 | ECO:0000255|PROSITE-ProRule:PRU00159 | 426 | ... | Q7Z698 | SUBUNIT: Forms a complex with MAPK3/ERK1 but n... | FUNCTION: Constitutively active serine/threoni... | ACTIVITY REGULATION: Constitutively activated ... | MLPFAPQDEPWDREMEVFSGGGASSGEVNGLKMVDEPMEEGEADSC... | 1 | 1 | 1 | 1 | 1 |
8 rows × 32 columns
pspa_no_TYR.uniprot.isin(human_active.Uniprot).value_counts()uniprot
True 368
False 13
Name: count, dtype: int64PSPA not in human uniprot active kd:
pspa_no_TYR[~pspa_no_TYR.uniprot.isin(human_active.Uniprot)]| uniprot | kinase | -5P | -5G | -5A | -5C | -5S | -5T | -5V | -5I | ... | 4E | 4s | 4t | 4y | 0s | 0t | 0y | 0S | 0T | 0Y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 8 | Q96QP1 | ALPHAK3 | 0.0571 | 0.0478 | 0.0253 | 0.0384 | 0.0571 | 0.0571 | 0.0586 | 0.0602 | ... | 0.0747 | 0.1142 | 0.1142 | 0.1744 | 0.1319 | 1.0000 | 0.0 | 0.1319 | 1.0000 | 0.0 |
| 18 | O14874 | BCKDK | 0.0482 | 0.0672 | 0.0598 | 0.0694 | 0.0566 | 0.0566 | 0.0517 | 0.0467 | ... | 0.0563 | 0.0401 | 0.0401 | 0.0859 | 1.0000 | 0.2348 | 0.0 | 1.0000 | 0.2348 | 0.0 |
| 26 | O43683 | BUB1 | 0.0899 | 0.0222 | 0.0249 | 0.0470 | 0.0286 | 0.0286 | 0.0319 | 0.0659 | ... | 0.0250 | 0.0261 | 0.0261 | 0.0273 | 0.5413 | 1.0000 | 0.0 | 0.5413 | 1.0000 | 0.0 |
| 59 | Q96QT4 | CHAK1 | 0.0649 | 0.0823 | 0.0686 | 0.0995 | 0.0608 | 0.0608 | 0.0431 | 0.0313 | ... | 0.0330 | 0.0400 | 0.0400 | 0.0318 | 1.0000 | 0.7003 | 0.0 | 1.0000 | 0.7003 | 0.0 |
| 60 | Q9BX84 | CHAK2 | 0.0532 | 0.0844 | 0.0761 | 0.0626 | 0.0588 | 0.0588 | 0.0439 | 0.0406 | ... | 0.0584 | 0.0610 | 0.0610 | 0.0538 | 1.0000 | 0.5794 | 0.0 | 1.0000 | 0.5794 | 0.0 |
| 93 | O00418 | EEF2K | 0.0603 | 0.0627 | 0.0635 | 0.0586 | 0.0602 | 0.0602 | 0.0584 | 0.0597 | ... | 0.0379 | 0.0620 | 0.0620 | 0.0434 | 0.2741 | 1.0000 | 0.0 | 0.2741 | 1.0000 | 0.0 |
| 98 | Q8IXL6 | FAM20C | 0.0496 | 0.0620 | 0.0669 | 0.0649 | 0.0564 | 0.0564 | 0.0564 | 0.0442 | ... | 0.1123 | 0.1801 | 0.1801 | 0.1610 | 1.0000 | 0.1276 | 0.0 | 1.0000 | 0.1276 | 0.0 |
| 111 | Q8TF76 | HASPIN | 0.0775 | 0.0522 | 0.0492 | 0.0495 | 0.0522 | 0.0522 | 0.0578 | 0.1577 | ... | 0.0237 | 0.0381 | 0.0381 | 0.0347 | 0.3998 | 1.0000 | 0.0 | 0.3998 | 1.0000 | 0.0 |
| 212 | Q15118 | PDHK1 | 0.0451 | 0.0697 | 0.0594 | 0.0625 | 0.0594 | 0.0594 | 0.0573 | 0.0590 | ... | 0.0535 | 0.0548 | 0.0548 | 0.0575 | 1.0000 | 0.4886 | 0.0 | 1.0000 | 0.4886 | 0.0 |
| 213 | Q16654 | PDHK4 | 0.0452 | 0.0645 | 0.0665 | 0.0672 | 0.0622 | 0.0622 | 0.0515 | 0.0619 | ... | 0.0608 | 0.0848 | 0.0848 | 0.0628 | 1.0000 | 0.4640 | 0.0 | 1.0000 | 0.4640 | 0.0 |
| 270 | Q96SB4 | SRPK1 | 0.0594 | 0.0753 | 0.0889 | 0.0814 | 0.0525 | 0.0525 | 0.0517 | 0.0468 | ... | 0.0521 | 0.0701 | 0.0701 | 0.0622 | 1.0000 | 0.2897 | 0.0 | 1.0000 | 0.2897 | 0.0 |
| 271 | P78362 | SRPK2 | 0.0446 | 0.0660 | 0.0596 | 0.0694 | 0.0491 | 0.0491 | 0.0452 | 0.0349 | ... | 0.0440 | 0.0562 | 0.0562 | 0.0559 | 1.0000 | 0.1949 | 0.0 | 1.0000 | 0.1949 | 0.0 |
| 272 | Q9UPE1 | SRPK3 | 0.0435 | 0.0618 | 0.0556 | 0.0622 | 0.0541 | 0.0541 | 0.0527 | 0.0541 | ... | 0.0394 | 0.0514 | 0.0514 | 0.0544 | 1.0000 | 0.4323 | 0.0 | 1.0000 | 0.4323 | 0.0 |
13 rows × 215 columns
pspa_active_kd = pspa_no_TYR[pspa_no_TYR.uniprot.isin(human_active.Uniprot)]# some category test _TYR and have double annotation, split by _
info['pspa_category'] = info.pspa_category_big.str.split('_').str[0]info['pspa_category_detail']= info.pspa_category_small.str.split('_').str[0]info = info[info.pseudo=='0']category = info[['uniprot','kinase','group','pspa_category','pspa_category_detail']]head = pd.DataFrame(pspa_active_kd.uniprot).merge(category)pspa_active_kd = pspa_active_kd.reset_index(drop=True)head| uniprot | kinase | group | pspa_category | pspa_category_detail | |
|---|---|---|---|---|---|
| 0 | Q2M2I8 | AAK1 | Other | NAK | NAK |
| 1 | P27037 | ACVR2A | TKL | acidophilic | TGFBR |
| 2 | Q13705 | ACVR2B | TKL | acidophilic | TGFBR |
| 3 | P31749 | AKT1 | AGC | basophilic | AKT/ROCK |
| 4 | P31751 | AKT2 | AGC | basophilic | AKT/ROCK |
| ... | ... | ... | ... | ... | ... |
| 363 | P17948 | FLT1 | TK | FGF and VEGF receptors | FGF and VEGF receptors |
| 364 | P35968 | KDR | TK | FGF and VEGF receptors | FGF and VEGF receptors |
| 365 | P35916 | FLT4 | TK | FGF and VEGF receptors | FGF and VEGF receptors |
| 366 | P07947 | YES1 | TK | SRC | SRC |
| 367 | P43403 | ZAP70 | TK | SYK and FAK | SYK and FAK |
368 rows × 5 columns
pspa_active_kd = pd.concat([head,pspa_active_kd.iloc[:,2:]],axis=1)# pspa_active_kd.to_csv('out/pspa_uniprot_unique_no_TYR_category.csv',index=False)pspa_active_kd| uniprot | kinase | group | pspa_category | pspa_category_detail | -5P | -5G | -5A | -5C | -5S | ... | 4E | 4s | 4t | 4y | 0s | 0t | 0y | 0S | 0T | 0Y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Q2M2I8 | AAK1 | Other | NAK | NAK | 0.0720 | 0.0245 | 0.0284 | 0.0456 | 0.0425 | ... | 0.0457 | 0.0251 | 0.0251 | 0.0270 | 0.1013 | 1.0000 | 0.0 | 0.1013 | 1.0000 | 0.0 |
| 1 | P27037 | ACVR2A | TKL | acidophilic | TGFBR | 0.0415 | 0.0481 | 0.0584 | 0.0489 | 0.0578 | ... | 0.0640 | 0.0703 | 0.0703 | 0.0589 | 0.9833 | 1.0000 | 0.0 | 0.9833 | 1.0000 | 0.0 |
| 2 | Q13705 | ACVR2B | TKL | acidophilic | TGFBR | 0.0533 | 0.0517 | 0.0566 | 0.0772 | 0.0533 | ... | 0.0697 | 0.0761 | 0.0761 | 0.0637 | 0.9593 | 1.0000 | 0.0 | 0.9593 | 1.0000 | 0.0 |
| 3 | P31749 | AKT1 | AGC | basophilic | AKT/ROCK | 0.0603 | 0.0594 | 0.0552 | 0.0605 | 0.0516 | ... | 0.0312 | 0.0393 | 0.0393 | 0.0263 | 1.0000 | 0.6440 | 0.0 | 1.0000 | 0.6440 | 0.0 |
| 4 | P31751 | AKT2 | AGC | basophilic | AKT/ROCK | 0.0602 | 0.0617 | 0.0643 | 0.0582 | 0.0534 | ... | 0.0350 | 0.0548 | 0.0548 | 0.0417 | 1.0000 | 0.6077 | 0.0 | 1.0000 | 0.6077 | 0.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 363 | P17948 | FLT1 | TK | FGF and VEGF receptors | FGF and VEGF receptors | 0.0642 | 0.0688 | 0.0597 | 0.0618 | 0.0614 | ... | 0.0510 | 0.0677 | 0.0677 | 0.0380 | 0.0000 | 0.0000 | 1.0 | 0.0000 | 0.0000 | 1.0 |
| 364 | P35968 | KDR | TK | FGF and VEGF receptors | FGF and VEGF receptors | 0.0634 | 0.0672 | 0.0556 | 0.0517 | 0.0541 | ... | 0.0338 | 0.0300 | 0.0300 | 0.0292 | 0.0000 | 0.0000 | 1.0 | 0.0000 | 0.0000 | 1.0 |
| 365 | P35916 | FLT4 | TK | FGF and VEGF receptors | FGF and VEGF receptors | 0.0457 | 0.0531 | 0.0488 | 0.0553 | 0.0512 | ... | 0.0497 | 0.0445 | 0.0445 | 0.0500 | 0.0000 | 0.0000 | 1.0 | 0.0000 | 0.0000 | 1.0 |
| 366 | P07947 | YES1 | TK | SRC | SRC | 0.0677 | 0.0571 | 0.0537 | 0.0530 | 0.0527 | ... | 0.0492 | 0.0371 | 0.0371 | 0.0467 | 0.0000 | 0.0000 | 1.0 | 0.0000 | 0.0000 | 1.0 |
| 367 | P43403 | ZAP70 | TK | SYK and FAK | SYK and FAK | 0.0602 | 0.0880 | 0.0623 | 0.0496 | 0.0471 | ... | 0.0558 | 0.0440 | 0.0440 | 0.0318 | 0.0000 | 0.0000 | 1.0 | 0.0000 | 0.0000 | 1.0 |
368 rows × 218 columns
id_map = human_active[['Uniprot','kd_ID']]
id_map.columns=['uniprot','kd_ID']active_id = id_map[id_map.uniprot.isin(pspa_active_kd.uniprot)]Uniprot in pspa with two kinase domains
pspa_2kd = active_id[active_id.uniprot.duplicated(keep=False)].sort_values('uniprot')pspa_2kd| uniprot | kd_ID | |
|---|---|---|
| 988 | O75582 | O75582_KS6A5_HUMAN_KD1 |
| 989 | O75582 | O75582_KS6A5_HUMAN_KD2 |
| 990 | O75676 | O75676_KS6A4_HUMAN_KD1 |
| 991 | O75676 | O75676_KS6A4_HUMAN_KD2 |
| 1918 | P51812 | P51812_KS6A3_HUMAN_KD1 |
| 1919 | P51812 | P51812_KS6A3_HUMAN_KD2 |
| 2560 | Q15349 | Q15349_KS6A2_HUMAN_KD1 |
| 2561 | Q15349 | Q15349_KS6A2_HUMAN_KD2 |
| 2563 | Q15418 | Q15418_KS6A1_HUMAN_KD1 |
| 2564 | Q15418 | Q15418_KS6A1_HUMAN_KD2 |
| 5338 | Q9UK32 | Q9UK32_KS6A6_HUMAN_KD1 |
| 5339 | Q9UK32 | Q9UK32_KS6A6_HUMAN_KD2 |
pspa_2kd.uniprot.unique()array(['O75582', 'O75676', 'P51812', 'Q15349', 'Q15418', 'Q9UK32'],
dtype=object)pspa_active_kd_remove2kd = pspa_active_kd[~pspa_active_kd.uniprot.isin(pspa_2kd.uniprot.unique())]pspa_active_kd_remove2kd = pspa_active_kd_remove2kd.reset_index(drop=True)head = pd.DataFrame(pspa_active_kd_remove2kd.uniprot).merge(id_map)head| uniprot | kd_ID | |
|---|---|---|
| 0 | Q2M2I8 | Q2M2I8_AAK1_HUMAN_KD1 |
| 1 | P27037 | P27037_AVR2A_HUMAN_KD1 |
| 2 | Q13705 | Q13705_AVR2B_HUMAN_KD1 |
| 3 | P31749 | P31749_AKT1_HUMAN_KD1 |
| 4 | P31751 | P31751_AKT2_HUMAN_KD1 |
| ... | ... | ... |
| 357 | P17948 | P17948_VGFR1_HUMAN_KD1 |
| 358 | P35968 | P35968_VGFR2_HUMAN_KD1 |
| 359 | P35916 | P35916_VGFR3_HUMAN_KD1 |
| 360 | P07947 | P07947_YES_HUMAN_KD1 |
| 361 | P43403 | P43403_ZAP70_HUMAN_KD1 |
362 rows × 2 columns
pspa_active_kd_remove2kd = pd.concat([head,pspa_active_kd_remove2kd.iloc[:,1:]],axis=1)# pspa_active_kd_remove2kd.to_csv('out/pspa_uniprot_unique_no_TYR_category_remove2kd.csv')pspa_active_kd_remove2kd| uniprot | kd_ID | kinase | group | pspa_category | pspa_category_detail | -5P | -5G | -5A | -5C | ... | 4E | 4s | 4t | 4y | 0s | 0t | 0y | 0S | 0T | 0Y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Q2M2I8 | Q2M2I8_AAK1_HUMAN_KD1 | AAK1 | Other | NAK | NAK | 0.0720 | 0.0245 | 0.0284 | 0.0456 | ... | 0.0457 | 0.0251 | 0.0251 | 0.0270 | 0.1013 | 1.0000 | 0.0 | 0.1013 | 1.0000 | 0.0 |
| 1 | P27037 | P27037_AVR2A_HUMAN_KD1 | ACVR2A | TKL | acidophilic | TGFBR | 0.0415 | 0.0481 | 0.0584 | 0.0489 | ... | 0.0640 | 0.0703 | 0.0703 | 0.0589 | 0.9833 | 1.0000 | 0.0 | 0.9833 | 1.0000 | 0.0 |
| 2 | Q13705 | Q13705_AVR2B_HUMAN_KD1 | ACVR2B | TKL | acidophilic | TGFBR | 0.0533 | 0.0517 | 0.0566 | 0.0772 | ... | 0.0697 | 0.0761 | 0.0761 | 0.0637 | 0.9593 | 1.0000 | 0.0 | 0.9593 | 1.0000 | 0.0 |
| 3 | P31749 | P31749_AKT1_HUMAN_KD1 | AKT1 | AGC | basophilic | AKT/ROCK | 0.0603 | 0.0594 | 0.0552 | 0.0605 | ... | 0.0312 | 0.0393 | 0.0393 | 0.0263 | 1.0000 | 0.6440 | 0.0 | 1.0000 | 0.6440 | 0.0 |
| 4 | P31751 | P31751_AKT2_HUMAN_KD1 | AKT2 | AGC | basophilic | AKT/ROCK | 0.0602 | 0.0617 | 0.0643 | 0.0582 | ... | 0.0350 | 0.0548 | 0.0548 | 0.0417 | 1.0000 | 0.6077 | 0.0 | 1.0000 | 0.6077 | 0.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 357 | P17948 | P17948_VGFR1_HUMAN_KD1 | FLT1 | TK | FGF and VEGF receptors | FGF and VEGF receptors | 0.0642 | 0.0688 | 0.0597 | 0.0618 | ... | 0.0510 | 0.0677 | 0.0677 | 0.0380 | 0.0000 | 0.0000 | 1.0 | 0.0000 | 0.0000 | 1.0 |
| 358 | P35968 | P35968_VGFR2_HUMAN_KD1 | KDR | TK | FGF and VEGF receptors | FGF and VEGF receptors | 0.0634 | 0.0672 | 0.0556 | 0.0517 | ... | 0.0338 | 0.0300 | 0.0300 | 0.0292 | 0.0000 | 0.0000 | 1.0 | 0.0000 | 0.0000 | 1.0 |
| 359 | P35916 | P35916_VGFR3_HUMAN_KD1 | FLT4 | TK | FGF and VEGF receptors | FGF and VEGF receptors | 0.0457 | 0.0531 | 0.0488 | 0.0553 | ... | 0.0497 | 0.0445 | 0.0445 | 0.0500 | 0.0000 | 0.0000 | 1.0 | 0.0000 | 0.0000 | 1.0 |
| 360 | P07947 | P07947_YES_HUMAN_KD1 | YES1 | TK | SRC | SRC | 0.0677 | 0.0571 | 0.0537 | 0.0530 | ... | 0.0492 | 0.0371 | 0.0371 | 0.0467 | 0.0000 | 0.0000 | 1.0 | 0.0000 | 0.0000 | 1.0 |
| 361 | P43403 | P43403_ZAP70_HUMAN_KD1 | ZAP70 | TK | SYK and FAK | SYK and FAK | 0.0602 | 0.0880 | 0.0623 | 0.0496 | ... | 0.0558 | 0.0440 | 0.0440 | 0.0318 | 0.0000 | 0.0000 | 1.0 | 0.0000 | 0.0000 | 1.0 |
362 rows × 219 columns
Onehot encode
freq_max| position | aa | max_value | |
|---|---|---|---|
| 0 | 1549 | N | 0.815390 |
| 1 | 2618 | D | 0.809429 |
| 2 | 1724 | D | 0.800759 |
| 3 | 1525 | D | 0.791004 |
| 4 | 1730 | G | 0.775470 |
| ... | ... | ... | ... |
| 3429 | 20 | G | 0.000181 |
| 3430 | 21 | I | 0.000181 |
| 3431 | 22 | D | 0.000181 |
| 3432 | 23 | A | 0.000181 |
| 3433 | 3363 | H | 0.000181 |
3434 rows × 3 columns
onehot_col = freq_max[freq_max.max_value>0.05].position.sort_values().tolist()onehot = df[onehot_col]onehot_pspa = onehot.loc[pspa_active_kd_remove2kd.kd_ID]onehot_pspa.shape(362, 373)from sklearn.preprocessing import OneHotEncoder
def get_onehot(df):
df=df.copy()
encoded_df = pd.DataFrame(index=df.index)
encoder = OneHotEncoder(sparse_output=False, dtype=int, handle_unknown='ignore')
for col in df.columns:
reshaped = df[[col]] # keep as DataFrame
encoded = encoder.fit_transform(reshaped)
aa_labels = encoder.categories_[0]
new_col_names = [f"{col}_{aa}" for aa in aa_labels]
encoded_subdf = pd.DataFrame(encoded,index=df.index)
encoded_subdf.columns=new_col_names
encoded_df = pd.concat([encoded_df, encoded_subdf], axis=1)
return encoded_dfencoded_all=get_onehot(onehot)encoded_all.shape(5536, 7567)encoded_all.head()| 65_- | 65_A | 65_C | 65_D | 65_E | 65_F | 65_G | 65_H | 65_I | 65_K | ... | 3192_M | 3192_N | 3192_P | 3192_Q | 3192_R | 3192_S | 3192_T | 3192_V | 3192_W | 3192_Y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kd_ID | |||||||||||||||||||||
| A0A075F7E9_LERK1_ORYSI_KD1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A0A078BQP2_GCY25_CAEEL_KD1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A0A078CGE6_M3KE1_BRANA_KD1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A0A0G2K344_PK3CA_RAT_KD1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| A0A0H2ZM62_HK06_STRP2_KD1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
5 rows × 7567 columns
from katlas.feature import *
from katlas.plot import *reduce_feature?Signature: reduce_feature( df: pandas.core.frame.DataFrame, method: str = 'pca', complexity: int = 20, n: int = 2, load: str = None, save: str = None, seed: int = 123, **kwargs, ) Docstring: Reduce the dimensionality given a dataframe of values File: ~/katlas/katlas/feature.py Type: function
umap2d_all = reduce_feature(encoded_all,
'umap',
complexity=15,
min_dist=0.7
)/home/zeus/miniconda3/envs/cloudspace/lib/python3.10/site-packages/sklearn/utils/deprecation.py:151: FutureWarning: 'force_all_finite' was renamed to 'ensure_all_finite' in 1.6 and will be removed in 1.8.
warnings.warn(
/home/zeus/miniconda3/envs/cloudspace/lib/python3.10/site-packages/umap/umap_.py:1952: UserWarning: n_jobs value 1 overridden to 1 by setting random_state. Use no seed for parallelism.
warn(set_sns()import seaborn as sns
from matplotlib import pyplot as pltdef plot_umap(embedding_df,box_title,hue=None,palette='tab20',alpha=0.7,**kwargs):
'Plot umap.'
x_col, y_col = embedding_df.columns
g = sns.relplot(data=embedding_df, x=x_col, y=y_col, hue=hue,palette=palette,alpha=alpha,**kwargs)
g._legend.set_title(box_title)
plt.xticks([])
plt.yticks([])hue_is_PSPA=umap2d_all.index.isin(pspa_active_kd_remove2kd.kd_ID)TF_palette = {True: 'red', False: 'gray'}plot_umap(umap2d_all,
box_title='Known PSPA',alpha=0.5,
hue = hue_is_PSPA,hue_order=[True, False],
s=9,
palette=TF_palette)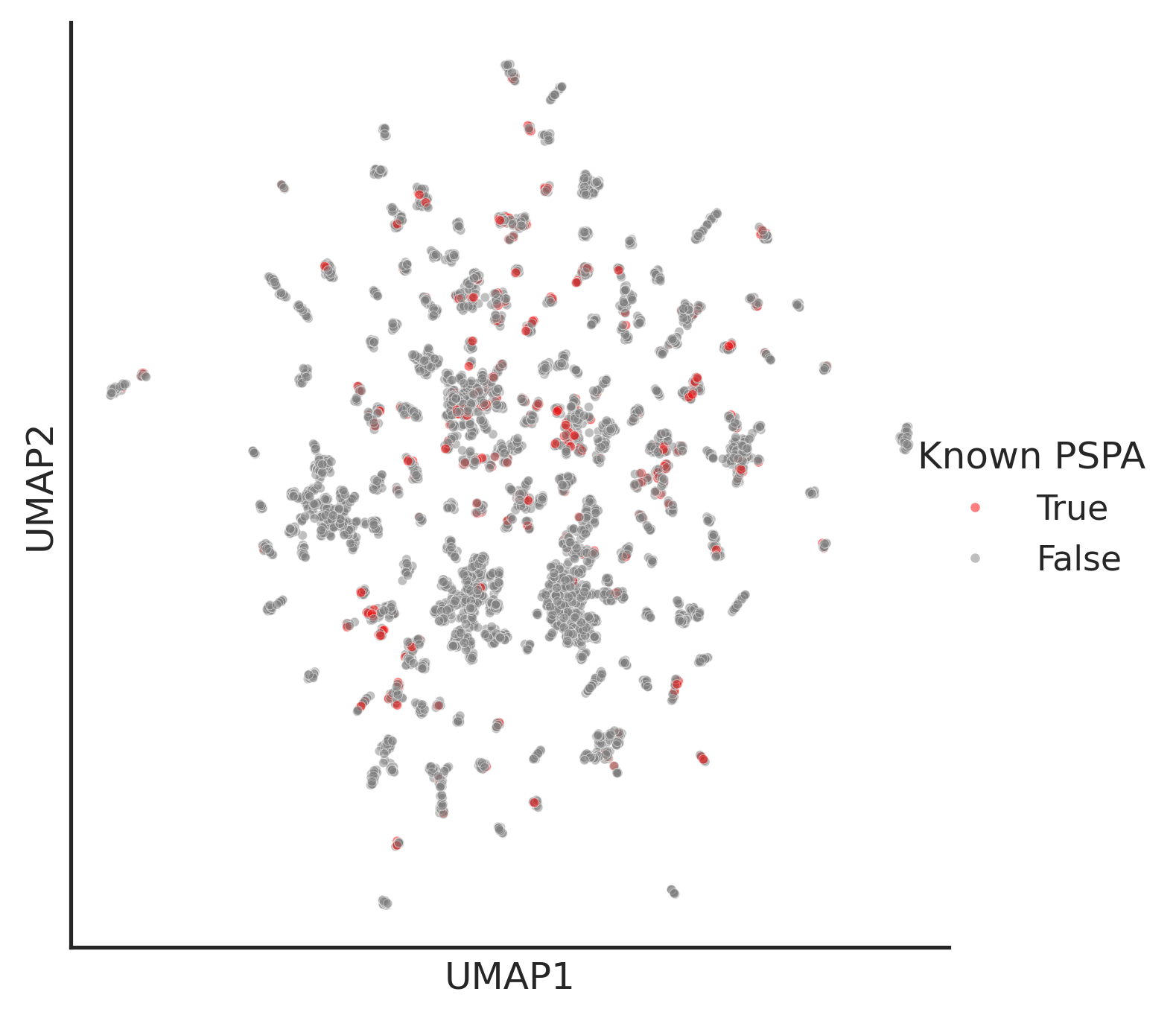
PSPA kd plot
encoded_df = get_onehot(onehot_pspa)encoded_df| 65_- | 65_A | 65_C | 65_E | 65_F | 65_I | 65_K | 65_L | 65_N | 65_Q | ... | 3192_K | 3192_L | 3192_M | 3192_Q | 3192_R | 3192_S | 3192_T | 3192_V | 3192_W | 3192_Y | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| kd_ID | |||||||||||||||||||||
| Q2M2I8_AAK1_HUMAN_KD1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| P27037_AVR2A_HUMAN_KD1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Q13705_AVR2B_HUMAN_KD1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| P31749_AKT1_HUMAN_KD1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| P31751_AKT2_HUMAN_KD1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| P17948_VGFR1_HUMAN_KD1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| P35968_VGFR2_HUMAN_KD1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| P35916_VGFR3_HUMAN_KD1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| P07947_YES_HUMAN_KD1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| P43403_ZAP70_HUMAN_KD1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
362 rows × 5121 columns
umap2d = reduce_feature(encoded_df,
'umap',
complexity=15,
min_dist=0.7
)/home/zeus/miniconda3/envs/cloudspace/lib/python3.10/site-packages/sklearn/utils/deprecation.py:151: FutureWarning: 'force_all_finite' was renamed to 'ensure_all_finite' in 1.6 and will be removed in 1.8.
warnings.warn(
/home/zeus/miniconda3/envs/cloudspace/lib/python3.10/site-packages/umap/umap_.py:1952: UserWarning: n_jobs value 1 overridden to 1 by setting random_state. Use no seed for parallelism.
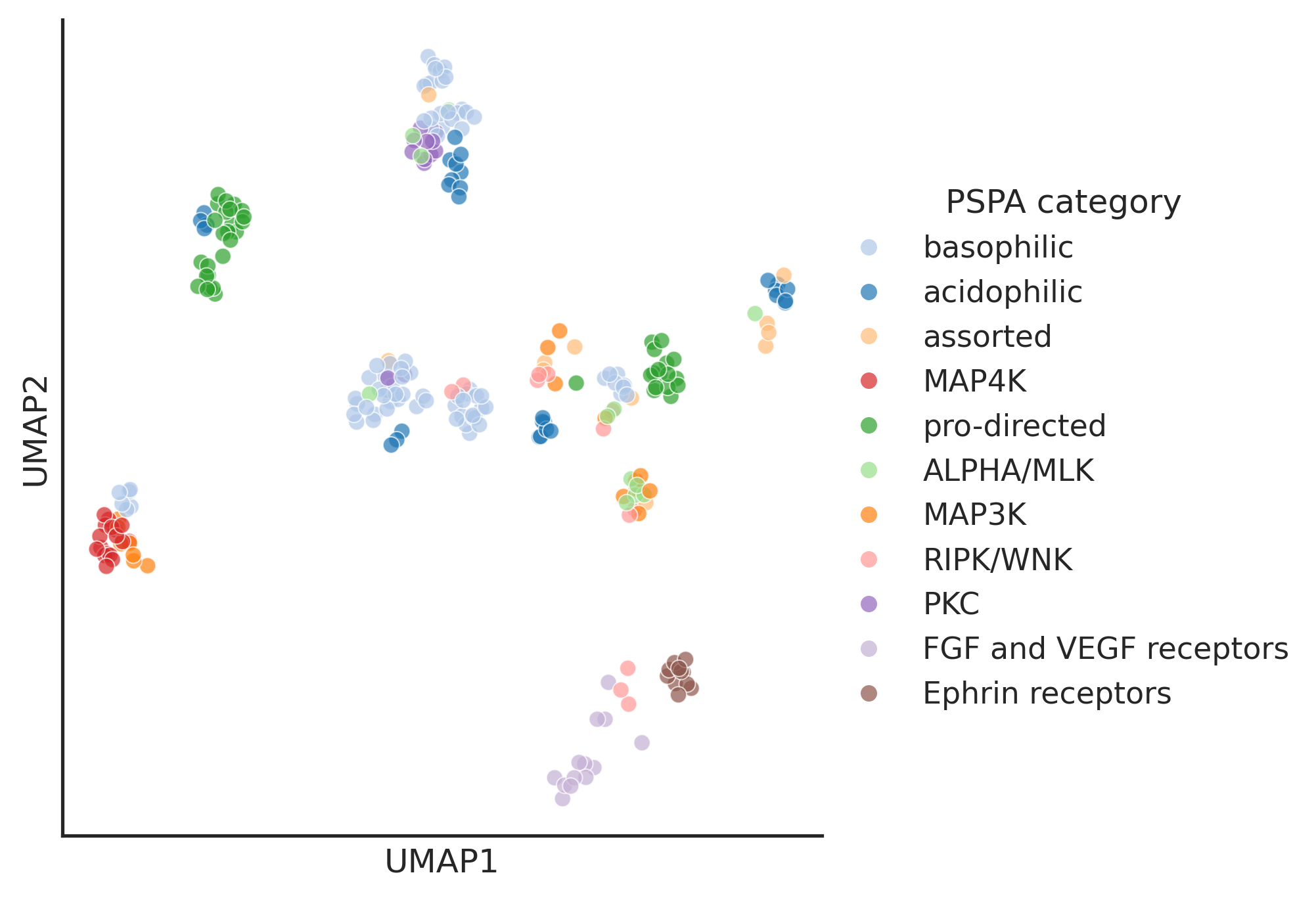
warn(hue_pspa = pspa_active_kd_remove2kd.set_index('kd_ID')['pspa_category']
hue_group = pspa_active_kd_remove2kd.set_index('kd_ID')['group']enrich_group = hue_pspa.value_counts()[hue_pspa.value_counts()>10].indexhue_pspa_partial = hue_pspa[hue_pspa.isin(enrich_group)]group_color = pd.read_csv('raw/group_color.csv')group_color = group_color.set_index('Label')['Color'].to_dict()plot_umap(umap2d,box_title='Group',hue=hue_group, palette=group_color)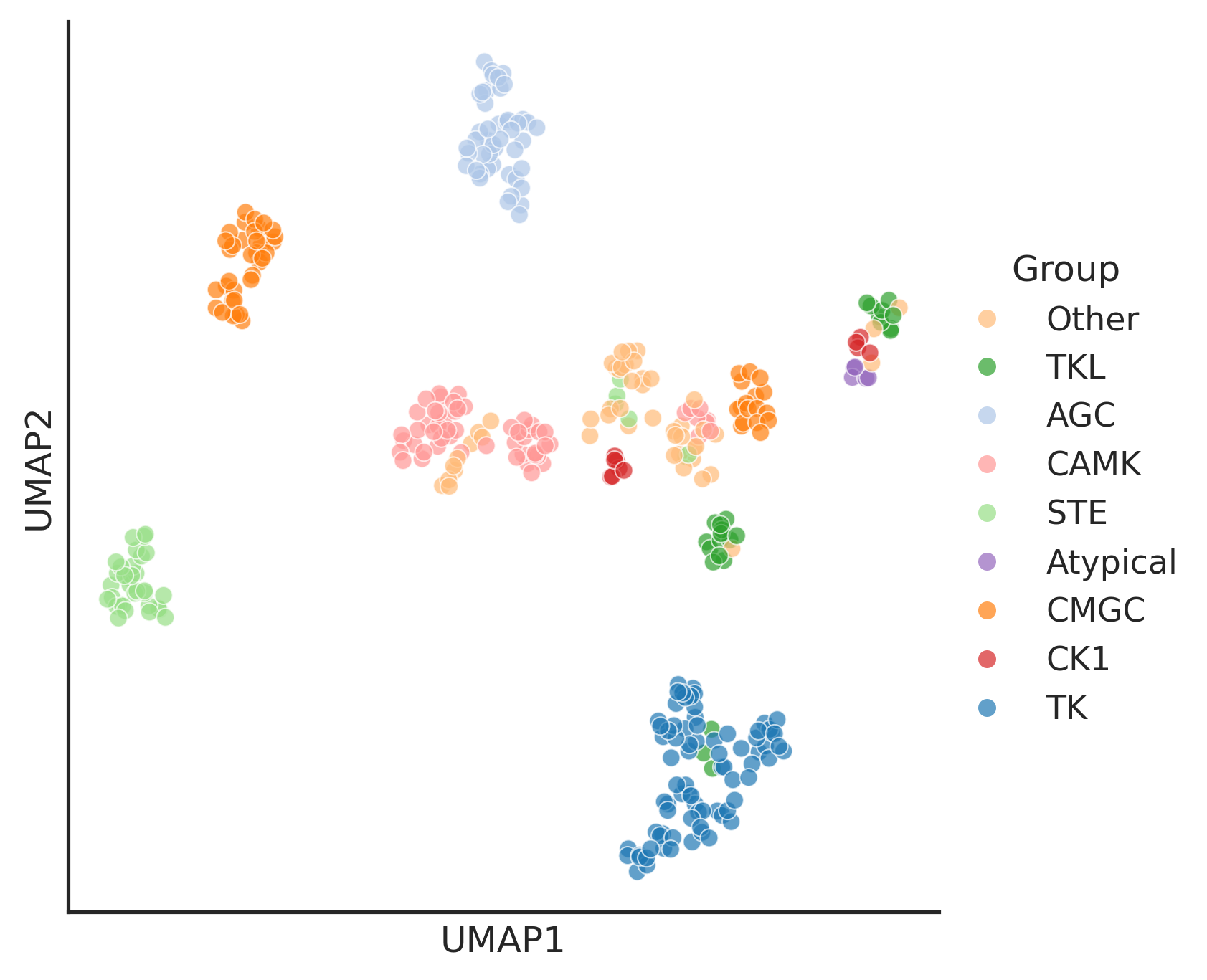
pspa_color = get_color_dict(hue_pspa_partial.unique())def rgb_to_hex(rgb_dict):
"""
Convert a dictionary with RGB values in [0, 1] to hex color codes.
"""
hex_dict = {
key: '#{:02X}{:02X}{:02X}'.format(
int(r * 255), int(g * 255), int(b * 255)
)
for key, (r, g, b) in rgb_dict.items()
}
return hex_dictpspa_color = rgb_to_hex(pspa_color)pspa_color = pd.DataFrame.from_dict(pspa_color,orient='index').reset_index()pspa_color.columns=['Label','Color']# pspa_color.to_csv('raw/pspa_color.csv',index=False)pspa_color=pd.read_csv('raw/pspa_color.csv')pspa_color = pspa_color.set_index('Label')['Color'].to_dict()plot_umap(umap2d,box_title='PSPA category',hue=hue_pspa_partial,palette=pspa_color)